Abstract
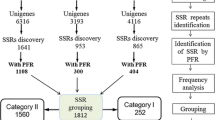
Due to their highly polymorphic and codominant nature, simple-sequence repeat (SSR) markers are a common choice for assaying genetic diversity and genetic mapping. In this paper, we describe the generation of an expressed-sequence tag (EST) collection for the oilseed crop safflower and the subsequent development of EST–SSR markers for the genetic analysis of safflower and related species. We assembled 40,874 reads into 19,395 unigenes, of which 4,416 (22.8%) contained at least one SSR. Primer pairs were developed and tested for 384 of these loci, resulting in a collection of 104 polymorphic markers that amplify reliably across 27 accessions (3 species) of the genus Carthamus. These markers exhibited a high level of polymorphism, with an average of 6.0 ± 0.4 alleles per locus and an average gene diversity of 0.54 ± 0.03 across Carthamus species. In terms of cross-taxon transferability, 50% of these primer pairs produced an amplicon in at least one other species in the Asteraceae, and 28% produced an amplicon in at least one species outside the safflower subfamily (i.e., lettuce, sunflower, and/or Gerbera). These markers represent a valuable resource for the genetic analysis of safflower and related species, and also have the potential to facilitate comparative map-based analyses across a broader array of taxa within the Asteraceae.


Similar content being viewed by others
References
Becher R (2007) EST-derived microsatellites as a rich source of molecular markers for oats. Plant Breed 126:274–278
Burke JM, Tang S, Knapp SJ, Rieseberg LH (2002) Genetic analysis of sunflower domestication. Genetics 161:1257–1267
Chapman MA, Burke JM (2007) DNA sequence diversity and the origin of cultivated safflower (Carthamus tinctorius L.; Asteraceae). BMC Plant Biol 7:60
Chapman MA, Chang J-C, Weisman D, Kesseli RV, Burke JM (2007) Universal markers for comparative mapping and phylogenetic analysis in the Asteraceae (Compositae). Theor Appl Genet 115:747–755
Chase MR, Moller C, Kesseli R, Bawa KS (1996) Distant gene flow in tropical trees. Nature 383:398–399
Cordeiro GM, Casu R, McIntyre CL, Manners JM, Henry RJ (2001) Microsatellite markers from sugarcane (Saccharum spp.) ESTs cross transferable to erianthus and sorghum. Plant Sci 160:1115–1123
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Ellis JR, Burke JM (2007) EST-SSRs as a resource for population genetic analyses. Heredity 99:125–132
Feingold S, Lloyd J, Norero N, Bonierbale M, Lorenzen J (2005) Mapping and characterization of new EST-derived microsatellites for potato (Solanum tuberosum L.). Theor Appl Genet 111:456–466
Gale MD, Devos KM (1998) Plant comparative genetics after ten years. Science 282:656–658
Gentzbittel L, Mestries E, Mouzeyar S, Mazeryat F, Badaoui S, Vear F, de la Tourvieille Brouhe D, Nicolas P (1999) A composite map of expressed sequences and phenotypic traits of the sunflower (Helianthus annuus L.) genome. Theor Appl Genet 99:218–234
Gupta M, Chyi Y-S, Romero-Stevenson J, Owen JL (1994) Amplification of DNA markers from evolutionarily diverse genomes using single primers of simple-sequence repeats. Theor Appl Genet 89:998–1006
Hanai LR, de Campos T, Camargo LEA, Benchimol LL, de Souza AP, Melotto M, Carbonell SAM, Chioratto AF, Consoli L, Formighieri EF, Siqueira M, Tsai SM, Vieira MLC (2007) Development, characterization, and comparative analysis of polymorphism at common bean SSR loci isolated from genic and genomic sources. Genome 50:266–277
Heesacker A, Kishore VK, Gao WX, Tang SX, Kolkman JM, Gingle A, Matvienko M, Kozik A, Michelmore RM, Lai Z, Rieseberg LH, Knapp SJ (2008) SSRs and INDELs mined from the sunflower EST database: abundance, polymorphisms, and cross-taxa utility. Theor Appl Genet 117:1021–1029
Johnson WC, Jackson LE, Ochoa O, van Wijk R, Peleman J, St. Clarir DA, Michelmore RW (2000) Lettuce, a shallow-rooted crop, and Lactuca serriola, its wild progenitor, differ at QTL determining root architecture and deep soil water exploitation. Theor Appl Genet 101:1066–1073
Johnson RC, Kisha TJ, Evans MA (2007) Characterizing safflower germplasm with AFLP molecular markers. Crop Sci 47:1728–1736
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Kesseli RV, Paran I, Michelmore RW (1994) Analysis of a detailed genetic linkage map of Lactuca sativa (Lettuce) constructed from RFLP and RAPD markers. Genetics 136:1435–1446
Knowles PF (1958) Safflower. Adv Agron 10:289–323
Kozik A, Michelmore RW, Knapp SJ, Matvienko MS, Rieseberg L, Lin H, van Damme M, Lavelle D, Chevalier P, Ziegle J, Ellison P, Kolkman J, Slaubaugh MB, Livingstone K, Zhou LZ, Church S, Edberg S, Jackson L, Bradford KJ (2002) Sunflower and lettuce ESTs developed by the Compositae Genome Project (http://compgenomics.ucdavis.edu/)
Lacey DJ, Wellner N, Beaudoin F, Napier JA, Shewry PR (1998) Secondary structure of oleosins in oil bodies isolated from seeds of safflower (Carthamus tinctorius L.) and sunflower (Helianthus annuus L.). Biochem J 334:469–477
Landry BS, Kesseli RV, Farrara B, Michelmore RW (1987) A genetic map of lettuce (Lactuca sativa L.) with restriction fragment length polymorphism, isozyme, disease resistance and morphological markers. Genetics 116:331–337
Laurent V, Devaux P, Thiel T, Viard F, Mielordt S, Touzet P, Quillet MC (2007) Comparative effectiveness of sugar beet microsatellite markers isolated from genomic libraries and GenBank ESTs to map the sugar beet genome. Theor Appl Genet 115:793–805
Levinson G, Gutman GA (1987) Slipped-strand mispairing—a major mechanism for DNA-sequence evolution. Mol Biol Evol 4:203–221
Lewis PO, Zaykin D (2001) Genetic data analysis: computer program for the analysis of allelic data. http://lewis.eeb.uconn.edu/lewishome/software.html
Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez GJ, Buckler E, Doebley J (2002) A single domestication for maize is shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA 99:6080–6084
McHale LK, Truco MJ, Kozik A, Lavelle DO, Ochoa OE, Wroblewski T, Knapp SJ, Michelmore RW (2009) Genomic architecture of disease resistance in lettuce. Theor Appl Genet 118:565–580
Neeraja CN, Maghirang-Rodriguez R, Pamplona A, Heuer S, Collard BCY, Septiningsih EM, Vergara G, Sanchez D, Xu K, Ismail AM, Mackill DJ (2007) A marker-assisted backcross approach for developing submergence-tolerant rice cultivars. Theor Appl Genet 115:767–776
Panero JL, Funk VA (2008) The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed. Mol Phylogenet Evol 47:757–782
Pashley CH, Ellis JR, McCauley DE, Burke JM (2006) EST databases as a source for molecular markers: lessons from Helianthus. J Hered 97:381–388
Paterson AH, Bowers JE, Burow MD, Draye X, Elsik CG, Jiang CX, Katsar CS, Lan TH, Lin YR, Ming RG, Wright RJ (2000) Comparative genomics of plant chromosomes. Plant Cell 12:1523–1539
Peakall R, Smouse PE (2002) GenAlEx V5.04: genetic analysis in excel. Population genetic software for teaching and research. Australian National University, Canberra, Australia. http://www.anu.edu.au/BoZo/GenAlEx/
Pinto LR, Oliveira KM, Ulian EC, Garcia AAF, de Souza AP (2004) Survey in the sugarcane expressed sequence tag database (SUCEST) for simple sequence repeats. Genome 47:795–804
Saha MC, Cooper JD, Mian MAR, Chekhovskiy K, May GD (2006) Tall fescue genomic SSR markers: development and transferability across multiple grass species. Theor Appl Genet 113:1449–1458
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnol 18:233–234
Sehgal D, Raina SN (2005) Genotyping safflower (Carthamus tinctorius) cultivars by DNA fingerprints. Euphytica 146:67–76
Simko I (2008) Development of EST-SSR markers for the study of population structure in lettuce (Lactuca sativa L.). J Hered. doi: 10.1093/jhered/esn072
Squirrell J, Hollingsworth PM, Woodhead M, Russell J, Lowe AJ, Gibby M, Powell W (2003) How much effort is required to isolate nuclear microsatellites from plants? Mol Ecol 12:1339–1348
Stevens PF (2006) Angiosperm phylogeny Website. Version 7. http://www.mobot.org/MOBOT/research/APweb
Susanna A, Garcia-Jacas N, Hidalgo O, Vilatersana R, Garnatje T (2006) The Cardueae (Compositae) revisited: Insights from ITS, trnL-trnF, and matK nuclear and chloroplast DNA analysis. Ann Mo Bot Gard 93:150–171
Tang S, Yu JK, Slabaugh MB, Shintani DK, Knapp SJ (2002) Simple sequence repeat map of the sunflower genome. Theor Appl Genet 105:1124–1136
Tang S, Kishore VK, Knapp SJ (2003) PCR-multiplexes for a genome-wide framework of simple sequence repeat marker loci in cultivated sunflower. Theor Appl Genet 107:6–19
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S (2001) Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res 11:1141–1452
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Truco MJ, Antonise R, Lavelle D, Ochoa O, Kozik A, Witsenboer H, Fort S, Jeuken MJW, Kesseli RV, Lindhout P, Michelmore RW, Peleman J (2007) A high-density, integrated genetic linkage map of lettuce (Lactuca spp.). Theor Appl Genet 115:735–746
Weiss EA (1971) Castor, sesame and safflower. Barnes and Noble, Inc, New York, NY
Wills DM, Burke JM (2007) QTL analysis of the early domestication of sunflower. Genetics 176:2589–2599
Wills DM, Hester ML, Liu A, Burke JM (2005) Chloroplast SSR polymorphisms in the Compositae and the mode of organellar inheritance in Helianthus annuus. Theor Appl Genet 110:941–947
Xiong LX, Liu KD, Dai XK, Xu CG, Zhang QF (1999) Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor Appl Genet 98:243–251
Yi GB, Lee JM, Lee S, Choi D, Kim BD (2006) Exploitation of pepper EST-SSRs and an SSR-based linkage map. Theor Appl Genet 114:113–130
Yu JK, Tang S, Slabaugh MB, Heesacker A, Cole G, Herring M, Soper J, Han F, Chu WC, Webb DM, Thompson L, Edwards KJ, Berry S, Leon AJ, Grondona M, Olungu C, Maes N, Knapp SJ (2003) Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci 43:367–387
Yu JK, Dake TM, Singh S, Benscher D, Li WL, Gill B, Sorrells ME (2004) Development and mapping of EST-derived simple sequence repeat markers for hexaploid wheat. Genome 47:805–818
Acknowledgments
We would like to thank the greenhouse staff at UGA for maintenance of the safflower plants, the JGI for sequencing, data processing and delivery of the EST data and Huaqin Xu for maintenance of the CGPDB website. This work was supported by grants to J.M.B. from the National Science Foundation Plant Genome Research Program (DBI-0332411) and the Plant Genome Program of the USDA Cooperative State Research, Education, and Extension Service-National Research Initiative (03-35300-13104), and to the Compositae Genome Project from the USDA Initiative for Future Agriculture and Food Systems (IFAFS 2000-04292) and the NSF Plant Genome Research Program (DBI-0421630).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Bervillé.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chapman, M.A., Hvala, J., Strever, J. et al. Development, polymorphism, and cross-taxon utility of EST–SSR markers from safflower (Carthamus tinctorius L.). Theor Appl Genet 120, 85–91 (2009). https://doi.org/10.1007/s00122-009-1161-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-009-1161-8