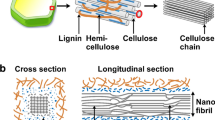
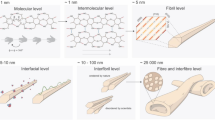
Abstract
Cellulose is produced by all plants and a number of other organisms, including bacteria. The most representative cellulose-producing bacterial species is Gluconacetobacter xylinus, an acetic acid bacterium. Cellulose produced by G. xylinus, commonly referred to as bacterial cellulose (BC), has exceptional physicochemical properties resulting in its use in a variety of applications. All cellulose-producing organisms that synthesize cellulose microfibrils have membrane-localized protein complexes (also called terminal complexes or TCs) that contain the enzyme cellulose synthase and other proteins. The bacterium G. xylinus is a prolific cellulose producer and a model organism for studies on cellulose biosynthesis. The widths of cellulose fibers produced by Gluconacetobacter are 50–100 nm, suggesting that cellulose-synthesizing complexes are nanomachines spinning a nanofiber. At least four different proteins (BcsA, BcsB, BcsC, and BcsD) are included in TC from Gluconacetobacter, and the proposed function of each is as follows: BcsA, synthesis of a glucan chain through glycosyl transfer from UDP-glucose; BcsB, complexes with BcsA for cellulose synthase activity; BcsC, formation of a pore in the outer membrane through which a glucan chain is extruded; BcsD, regulates aggregation of glucan chains through four tunnel-like structures. In this review, we discuss structures and functions of these four and a few other proteins that have a role in cellulose biosynthesis in bacteria.
Graphic abstract




reproduced from Ross et al. 1991, and is used with permission of American Society for Microbiology


















Similar content being viewed by others
References
Abidi W, Zouhir S, Caleechurn M et al (2021) Architecture and regulation of an enterobacterial cellulose secretion system. Sci Adv 7:1–16. https://doi.org/10.1126/sciadv.abd8049
Acheson JF, Derewenda ZS, Zimmer J (2019) Architecture of the cellulose synthase outer membrane channel and its association with the periplasmic TPR domain. Structure 27:1855-1861.e3. https://doi.org/10.1016/j.str.2019.09.008
Acheson JF, Ho R, Goularte NF et al (2021) Molecular organization of the E. coli cellulose synthase macrocomplex. Nat Struct Mol Biol 28:310–318. https://doi.org/10.1038/s41594-021-00569-7
Alzari PM, Souchon H, Dominguez R (1996) The crystal structure of endoglucanase CelA, a family 8 glycosyl hydrolase from Clostridium thermocellum. Structure 4:265–275. https://doi.org/10.1016/S0969-2126(96)00031-7
Benziman M, Haigler CH, Brown RM (1980) Cellulose biogenesis: polymerization and crystallization are coupled processes in Acetobacter xylinum. Proc Natl Acad Sci U S A 77:6678–6682. https://doi.org/10.1073/pnas.77.11.6678
Brown AJ (1886) XLIII.—On an acetic ferment which forms cellulose. J Chem Soc, Trans 49:432–439. https://doi.org/10.1039/CT8864900432
Brown RM (1996) The biosynthesis of cellulose. J Macromol Sci Part A 33:1345–1373. https://doi.org/10.1080/10601329608014912
Brown RM, Willison JHM, Richardson CL (1976) Cellulose biosynthesis in Acetobacter xylinum: visualization of the site of synthesis and direct measurement of the in vivo process. Proc Natl Acad Sci U S A 73:4565–4569. https://doi.org/10.1073/pnas.73.12.4565
Castro C, Cleenwerck I, Trček J et al (2013) Gluconacetobacter medellinensis sp. nov., cellulose- and non-cellulose-producing acetic acid bacteria isolated from vinegar. Int J Syst Evol Microbiol 63:1119–1125. https://doi.org/10.1099/ijs.0.043414-0
Chen HP, Brown RM (1996) Immunochemical studies of the cellulose synthase complex in Acetobacter xylinum. Cellulose 3:63–75. https://doi.org/10.1007/bf02228791
D’Andrea LD, Lynne R (2003) TPR proteins: the versatile helix. Trends Biochem Sci 28:655–662. https://doi.org/10.1016/j.tibs.2003.10.007
Du J, Vepachedu V, Cho SH et al (2016) Structure of the cellulose synthase complex of Gluconacetobacter hansenii at 23.4 Å resolution. PLoS ONE 11:1–24. https://doi.org/10.1371/journal.pone.0155886
Fang J, Kawano S, Tajima K, Kondo T (2015) In vivo curdlan/cellulose bionanocomposite synthesis by genetically modified Gluconacetobacter xylinus. Biomacromol 16:3154–3160. https://doi.org/10.1021/acs.biomac.5b01075
Florea M, Hagemann H, Santosa G et al (2016) Engineering control of bacterial cellulose production using a genetic toolkit and a new cellulose producing strain. Proc Natl Acad Sci U S A 113:E3431–E3440. https://doi.org/10.1073/pnas.1522985113
Fontana JD, De Souza AM, Fontana CK et al (1990) Acetobacter cellulose pellicle as a temporary skin substitute. Appl Biochem Biotechnol 24–25:253–264. https://doi.org/10.1007/BF02920250
Fujiwara T, Komoda K, Sakurai N et al (2013) The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A. Biochem Biophys Res Commun 431:802–807. https://doi.org/10.1016/j.bbrc.2012.12.103
Gao M, Li J, Bao Z et al (2019) A natural in situ fabrication method of functional bacterial cellulose using a microorganism. Nat Commun 10:1–10. https://doi.org/10.1038/s41467-018-07879-3
Haigler CH, Chanzy H (1988) Electron diffraction analysis of the altered cellulose synthesized by Acetobacter xylinum in the presence of fluorescent brightening agents and direct dyes. J Ultrastruct Res Mol Struct Res 98:299–311. https://doi.org/10.1016/S0889-1605(88)80922-4
Haigler CH, Brown RM, Benziman M (1980) Calcofluor white ST alters the in vivo assembly of cellulose microfibrils. Science. https://doi.org/10.1126/science.7434003
Hon DNS (1994) Cellulose: a random walk along its historical path. Cellulose 1:1–25. https://doi.org/10.1007/BF00818796
Hu S-Q, Gao Y-G, Tajima K et al (2010) Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. Proc Natl Acad Sci 107:17957–17961. https://doi.org/10.1073/pnas.1000601107
Hyun Min K, Sung Hee S, Yu Ryang P, Yu Sam K (1998) Evidence that a β-1,4-endoglucanase secreted by Acetobacter xylinum plays an essential role for the formation of cellulose fiber. Biosci Biotechnol Biochem 62:2257–2259. https://doi.org/10.1271/bbb.62.2257
Ifuku S, Nogi M, Abe K et al (2007) Surface modification of bacterial cellulose nanofibers for property enhancement of optically transparent composites: dependence on acetyl-group DS. Biomacromol 8:1973–1978. https://doi.org/10.1021/bm070113b
Iyer PR, Catchmark J, Brown NR, Tien M (2011) Biochemical localization of a protein involved in synthesis of Gluconacetobacter hansenii cellulose. Cellulose 18:739–747. https://doi.org/10.1007/s10570-011-9504-4
Kawano S, Tajima K, Kono H et al (2002a) Effects of endogenous endo-β-1,4-glucanase on cellulose biosynthesis in Acetobacter xylinum ATCC23769. J Biosci Bioeng 94:275–281. https://doi.org/10.1016/S1389-1723(02)80162-1
Kawano S, Tajima K, Uemori Y et al (2002b) Cloning of cellulose synthesis related genes from Acetobacter xylinum ATCC23769 and ATCC53582: Comparison of cellulose synthetic ability between strains. DNA Res 9:149–156. https://doi.org/10.1093/dnares/9.5.149
Kawano S, Tajima K, Kono H et al (2008) Regulation of endoglucanase gene (cmcax) expression in Acetobacter xylinum. J Biosci Bioeng 106:88–94. https://doi.org/10.1263/jbb.106.88
Kawano Y, Saotome T, Ochiai Y et al (2011) Cellulose accumulation and a cellulose synthase gene are responsible for cell aggregation in the Cyanobacterium Thermosynechococcus vulcanus RKN. Plant Cell Physiol 52:957–966. https://doi.org/10.1093/pcp/pcr047
Kimura S, Chen HP, Saxena IM et al (2001) Localization of c-di-GMP-binding protein with the linear terminal complexes of Acetobacter xylinum. J Bacteriol 183:5668–5674. https://doi.org/10.1128/JB.183.19.5668-5674.2001
Klemm D, Schumann D, Udhardt U, Marsch S (2001) Bacterial synthesized cellulose - artificial blood vessels for microsurgery. Prog Polym Sci 26:1561–1603. https://doi.org/10.1016/S0079-6700(01)00021-1
Knott BC, Crowley MF, Himmel ME et al (2016) Simulations of cellulose translocation in the bacterial cellulose synthase suggest a regulatory mechanism for the dimeric structure of cellulose. Chem Sci 7:3108–3116. https://doi.org/10.1039/C5SC04558D
Kose R, Sunagawa N, Yoshida M, Tajima K (2013) One-step production of nanofibrillated bacterial cellulose (NFBC) from waste glycerol using Gluconacetobacter intermedius NEDO-01. Cellulose 20:2971–2979. https://doi.org/10.1007/s10570-013-0050-0
Koyama M, Helbert W, Imai T et al (1997) Parallel-up structure evidences the molecular directionality during biosynthesis of bacterial cellulose. Proc Natl Acad Sci U S A 94:9091–9095. https://doi.org/10.1073/pnas.94.17.9091
Kumagai A, Mizuno M, Kato N et al (2011) Ultrafine cellulose fibers produced by Asaia bogorensis, an acetic acid bacterium. Biomacromol 12:2815–2821. https://doi.org/10.1021/bm2005615
Lee JW, Deng F, Yeomans WG et al (2001) Direct Incorporation of glucosamine and N-acetylglucosamine into exopolymers by Gluconacetobacter xylinus (=Acetobacter xylinum) ATCC 10245: production of chitosan-cellulose and chitin-cellulose exopolymers. Appl Environ Microbiol 67:3970–3975. https://doi.org/10.1128/AEM.67.9.3970-3975.2001
Lin FC, Brown RM Jr (1989) Purification of cellulose synthase from Acetobacter xylinum. In: Schuerc C (ed) Cellulose and wood—chemistry and technology. John Wiley & Sons, New York, pp 473–492
Lin FC, Brown RM, Drake RR, Haley BE (1990) Identification of the uridine 5’-diphosphoglucose (UDP-Glc) binding subunit of cellulose synthase in Acetobacter xylinum using the photoaffinity probe 5-azido-UDP-Glc. J Biol Chem 265:4782–4784. https://doi.org/10.1016/s0021-9258(19)34039-6
Luo H, Xiong G, Huang Y et al (2008) Preparation and characterization of a novel COL/BC composite for potential tissue engineering scaffolds. Mater Chem Phys 110:193–196. https://doi.org/10.1016/j.matchemphys.2008.01.040
Matthysse AG, Holmes KV, Gurlitz RHG (1981) Elaboration of cellulose fibrils by Agrobacterium tumefaciens during attachment to carrot cells. J Bacteriol 145:583–595. https://doi.org/10.1128/jb.145.1.583-595.1981
Mazur O, Zimmer J (2011) Apo- and cellopentaose-bound structures of the bacterial cellulose synthase subunit BcsZ. J Biol Chem 286:17601–17606. https://doi.org/10.1074/jbc.M111.227660
Mehta K, Pfeffer S, Brown RM (2015) Characterization of an acsD disruption mutant provides additional evidence for the hierarchical cell-directed self-assembly of cellulose in Gluconacetobacter xylinus. Cellulose 22:119–137. https://doi.org/10.1007/s10570-014-0521-y
Morgan JLW, McNamara JT, Fischer M et al (2016) Observing cellulose biosynthesis and membrane translocation in crystallo. Nature 531:329–334. https://doi.org/10.1038/nature16966
Morgan JLW, McNamara JT, Zimmer J (2014) Mechanism of activation of bacterial cellulose synthase by cyclic di-GMP. Nat Struct Mol Biol 21:489–496. https://doi.org/10.1038/nsmb.2803
Morgan JLW, Strumillo J, Zimmer J (2013) Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493:181–186. https://doi.org/10.1038/nature11744
Nakai T, Moriya A, Tonouchi N et al (1998) Control of expression by the cellulose synthase (bcsA) promoter region from Acetobacter xylinum BPR 2001. Gene 213:93–100. https://doi.org/10.1016/S0378-1119(98)00191-7
Nakai T, Sugano Y, Shoda M et al (2013) Formation of highly twisted ribbons in a carboxymethylcellulase gene-disrupted strain of a cellulose-producing bacterium. J Bacteriol 195:958–964. https://doi.org/10.1128/JB.01473-12
Napoli C, Dazzo F, Hubbell D (1975) Production of cellulose microfibrils by Rhizobium. Appl Microbiol 30:123–131. https://doi.org/10.1128/aem.30.1.123-131.1975
Nicolas WJ, Ghosal D, Tocheva EI et al (2021) Structure of the bacterial cellulose ribbon and its assembly-guiding cytoskeleton by electron cryotomography. J Bacteriol. https://doi.org/10.1128/JB.00371-20
Nishi Y, Uryu M, Yamanaka S et al (1990) The structure and mechanical properties of sheets prepared from bacterial cellulose - Part 2 Improvement of the mechanical properties of sheets and their applicability to diaphragms of electroacoustic transducers. J Mater Sci 25:2997–3001. https://doi.org/10.1007/BF00584917
Nobles DR, Romanovicz DK, Brown RM (2001) Cellulose in Cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol 127:529–542. https://doi.org/10.1104/pp.010557
Nogi M, Abe K, Handa K et al (2006a) Property enhancement of optically transparent bionanofiber composites by acetylation. Appl Phys Lett 89:233123. https://doi.org/10.1063/1.2403901
Nogi M, Ifuku S, Abe K et al (2006b) Fiber-content dependency of the optical transparency and thermal expansion of bacterial nanofiber reinforced composites. Appl Phys Lett 88:133124. https://doi.org/10.1063/1.2191667
Nojima S, Fujishima A, Kato K et al (2017) Crystal structure of the flexible tandem repeat domain of bacterial cellulose synthesis subunit C. Sci Rep. https://doi.org/10.1038/s41598-017-12530-0
Omadjela O, Narahari A, Strumillo J et al (2013) BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis. Proc Natl Acad Sci 110:17856–17861. https://doi.org/10.1073/pnas.1314063110
Orelma H, Morales LO, Johansson L-S et al (2014) Affibody conjugation onto bacterial cellulose tubes and bioseparation of human serum albumin. RSC Adv 4:51440–51450. https://doi.org/10.1039/C4RA08882D
Otsuka M, Fujiwara K, Sugai Y et al (2004) Characterization of a water-insoluble polymer producing bacterium Enterobacter sp. CJF-002 for MEOR. J Japan Pet Inst 47:282–292. https://doi.org/10.1627/jpi.47.282
Penttilä PA, Imai T, Capron M et al (2018) Multimethod approach to understand the assembly of cellulose fibrils in the biosynthesis of bacterial cellulose. Cellulose 25:2771–2783. https://doi.org/10.1007/s10570-018-1755-x
Perotti GF, Barud HS, Messaddeq Y et al (2011) Bacterial cellulose-laponite clay nanocomposites. Polymer (guildf) 52:157–163. https://doi.org/10.1016/j.polymer.2010.10.062
Portela R, Leal CR, Almeida PL, Sobral RG (2019) Bacterial cellulose: a versatile biopolymer for wound dressing applications. Microb Biotechnol 12:586–610. https://doi.org/10.1111/1751-7915.13392
Römling U, Galperin MY (2015) Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol 23:545–557. https://doi.org/10.1016/j.tim.2015.05.005
Ross P, Mayer R, Benziman M (1991) Cellulose biosynthesis and function in bacteria. Microbiol Rev 55:35–58. https://doi.org/10.1128/mr.55.1.35-58.1991
Saxena IM, Lin FC, Brown RM (1990) Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol Biol 15:673–683. https://doi.org/10.1007/BF00016118
Saxena IM, Kudlicka K, Okuda K, Brown RM (1994) Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bacteriol 176:5735–5752. https://doi.org/10.1128/jb.176.18.5735-5752.1994
Saxena IM, Brown RM, Fevre M et al (1995) Multidomain architecture of beta-glycosyl transferases: implications for mechanism of action. J Bacteriol 177:1419–1424. https://doi.org/10.1128/JB.177.6.1419-1424.1995
Schmidt A, Gonzalez A, Morris RJ et al (2002) Advantages of high-resolution phasing: MAD to atomic resolution. Acta Crystallogr Sect D Biol Crystallogr 58:1433–1441. https://doi.org/10.1107/S0907444902011368
Shah J, Malcolm Brown R (2005) Towards electronic paper displays made from microbial cellulose. Appl Microbiol Biotechnol 66:352–355. https://doi.org/10.1007/s00253-004-1756-6
Standal R, Iversen TG, Coucheron DH et al (1994) A new gene required for cellulose production and a gene encoding cellulolytic activity in Acetobacter xylinum are colocalized with the bcs operon. J Bacteriol 176:665–672. https://doi.org/10.1128/jb.176.3.665-672.1994
Sun S, Imai T, Sugiyama J, Kimura S (2017) CesA protein is included in the terminal complex of Acetobacter. Cellulose 24:2017–2027. https://doi.org/10.1007/s10570-017-1237-6
Sunagawa N, Tajima K, Hosoda M et al (2012) Cellulose production by Enterobacter sp. CJF-002 and identification of genes for cellulose biosynthesis. Cellulose 19:1989–2001. https://doi.org/10.1007/s10570-012-9777-2
Sunagawa N, Fujiwara T, Yoda T et al (2013) Cellulose complementing factor (Ccp) is a new member of the cellulose synthase complex (terminal complex) in Acetobacter xylinum. J Biosci Bioeng 115:607–612. https://doi.org/10.1016/j.jbiosc.2012.12.021
Tahara N, Tonouchi N, Yano H, Yoshinaga F (1998a) Purification and characterization of exo-1,4-β-glucosidase from Acetobacter xylinum BPR2001. J Ferment Bioeng 85:589–594. https://doi.org/10.1016/S0922-338X(98)80010-X
Tahara N, Yano H, Yoshinaga F (1998b) Subsite structure of exo-1,4-β-glucosidase from Acetobacter xylinum BPR2001. J Ferment Bioeng 85:595–597. https://doi.org/10.1016/S0922-338X(98)80011-1
Tajima K, Nakajima K, Yamashita H et al (2001) Cloning and sequencing of the beta-glucosidase gene from Acetobacter xylinum ATCC 23769. DNA Res 8:263–269. https://doi.org/10.1093/dnares/8.6.263
Tajima K, Kusumoto R, Kose R et al (2017) One-step production of amphiphilic nanofibrillated cellulose using a cellulose-producing bacterium. Biomacromol 18:3432–3438. https://doi.org/10.1021/acs.biomac.7b01100
Teh MY, Ooi KH, Danny Teo SX et al (2019) An expanded synthetic biology toolkit for gene expression control in Acetobacteraceae. ACS Synth Biol 8:708–723. https://doi.org/10.1021/acssynbio.8b00168
Thongsomboon W, Serra DO, Possling A et al (2018) Phosphoethanolamine cellulose: a naturally produced chemically modified cellulose. Science. https://doi.org/10.1126/science.aao4096
Tonouchi N, Tahara N, Tsuchida T et al (1995) Addition of a small amount of an endoglucanase enhances cellulose production by Acetobacter xylinum. Biosci Biotechnol Biochem 59:805–808. https://doi.org/10.1271/bbb.59.805
Tonouchi N, Tahara N, Kojima Y et al (1997) A beta-glucosidase gene downstream of the cellulose synthase operon in cellulose-producing Acetobacter. Biosci Biotechnol Biochem 61:1789–1790. https://doi.org/10.1271/bbb.61.1789
Umeda Y, Hirano A, Ishibashi M et al (1999) Cloning of cellulose synthase genes from Acetobacter xylinum JCM 7664: Implication of a novel set of cellulose synthase genes. DNA Res 6:109–115. https://doi.org/10.1093/dnares/6.2.109
Uto T, Ikeda Y, Sunagawa N et al (2021) Molecular dynamics simulation of cellulose synthase subunit D octamer with cellulose chains from acetic acid bacteria: insight into dynamic behaviors and thermodynamics on substrate recognition. J Chem Theory Comput 17:488–496. https://doi.org/10.1021/acs.jctc.0c01027
Wang X, Kong D, Zhang Y et al (2016) All-biomaterial supercapacitor derived from bacterial cellulose. Nanoscale 8:9146–9150. https://doi.org/10.1039/C6NR01485B
Whitney JC, Howell PL (2013) Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol 21:63–72. https://doi.org/10.1016/j.tim.2012.10.001
Wong HC, Fear AL, Calhoon RD et al (1990) Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc Natl Acad Sci U S A 87:8130–8134. https://doi.org/10.1073/pnas.87.20.8130
Yadav V, Paniliatis BJ, Shi H et al (2010) Novel in vivo-degradable cellulose-chitin copolymer from metabolically engineered Gluconacetobacter xylinus. Appl Environ Microbiol 76:6257–6265. https://doi.org/10.1128/AEM.00698-10
Yamada Y, Yukphan P (2008) Genera and species in acetic acid bacteria. Int J Food Microbiol 125:15–24. https://doi.org/10.1016/j.ijfoodmicro.2007.11.077
Yamada Y, Yukphan P, Vu HTL et al (2012) Description of Komagataeibacter gen. nov., with proposals of new combinations (Acetobacteraceae). J Gen Appl Microbiol 58:397–404. https://doi.org/10.2323/jgam.58.397
Yano H, Sugiyama J, Nakagaito AN et al (2005) Optically Transparent Composites Reinforced with Networks of Bacterial Nanofibers. Adv Mater 17:153–155. https://doi.org/10.1002/adma.200400597
Yasutake Y, Kawano S, Tajima K et al (2006) Structural characterization of the Acetobacter xylinum endo-β-1,4-glucanase CMCax required for cellulose biosynthesis. Proteins Struct Funct Bioinforma 64:1069–1077. https://doi.org/10.1002/prot.21052
Zaar K (1979) The Gram-negative bacterium Acetobacter xylinum assembles a cellulose ribbon composed of a number of microfibrils in the longitudinal axis of its envelope. The zone of ribbon assembly was investigated by freeze-etch electron microscopy. Freeze-Etching R J Cell Biol 80:773–777
Zang S, Zhang R, Chen H et al (2015) Investigation on artificial blood vessels prepared from bacterial cellulose. Mater Sci Eng C 46:111–117. https://doi.org/10.1016/j.msec.2014.10.023
Acknowledgments
This work was financially supported by a JSPS Grant-in-Aid for Scientific Research (A) (No. 19H00950, T.I.) and (B) (No. 19H02549, K.T.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human/animal rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tajima, K., Imai, T., Yui, T. et al. Cellulose-synthesizing machinery in bacteria. Cellulose 29, 2755–2777 (2022). https://doi.org/10.1007/s10570-021-04225-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10570-021-04225-7