Abstract
With the availability of the whole genome sequence of Escherichia coli or Corynebacterium glutamicum, strategies for directed DNA manipulation have developed rapidly. DNA manipulation plays an important role in understanding the function of genes and in constructing novel engineering bacteria according to requirement. DNA manipulation involves modifying the autologous genes and expressing the heterogenous genes. Two alternative approaches, using electroporation linear DNA or recombinant suicide plasmid, allow a wide variety of DNA manipulation. However, the over-expression of the desired gene is generally executed via plasmid-mediation. The current review summarizes the common strategies used for genetically modifying E. coli and C. glutamicum genomes, and discusses the technical problem of multi-layered DNA manipulation. Strategies for gene over-expression via integrating into genome are proposed. This review is intended to be an accessible introduction to DNA manipulation within the bacterial genome for novices and a source of the latest experimental information for experienced investigators.
中文概要
概 要
该综述较为全面地概述了当前针对大肠杆菌和谷 氨酸棒杆菌基因组遗传改造的各个方法的具体 流程、应用范围、注意事项以及其新颖之处, 比 较了针对基因定点突变、基因失活和基因过表达 的各个方法所存在的优缺点, 同时简单地介绍了 利用质粒介导基因过表达所存在的问题。此外, 还介绍了四种引物设计软件, 并简单分析了它们 的应用范围。为拟计划开展分子生物学实验的新 手对关于细菌基因组遗传改造方法做了可靠的 介绍, 同时也为已进行相关实验的实验员提供关 于基因定点突变、基因失活和基因过表达的最新 信息。
Similar content being viewed by others
1 Introduction
It is now well established that bacteria, such as Escherichia coli, Corynebacterium glutamicum, and yeast, exhibit excellent characteristics in producing amino acids, organic acids, and vitamins, etc. (Hou et al., 2012; Xu et al., 2013; 2014a; 2014b; 2015). Although the high-producing strains were obtained via repeated physical and/or chemical mutagenesis, there are many disadvantages to the processes, such as slow-growth, low sugar consumption rating, and low stress tolerance (Xu et al., 2013). Fortunately, the development of techniques for directed DNA manipulation and the availability of whole genome sequences make real the possibility of constructing high-producing strains using genetic engineering techniques (Kalinowski et al., 2003; Baba et al., 2006). These strategies have been successfully used in modifying E. coli or C. glutamicum strains for producing, e.g., amino acids (Georgi et al., 2005; Becker et al., 2011), organic acids (Inui et al., 2007), and vitamins (Martens et al., 2002). The strategies of genetic engineering breeding for constructing high-producing strains are as follows: (1) up-regulation of the key enzyme gene involved in the target product biosynthetic pathways; (2) relieving the inhibition and/or repression of the key enzyme; (3) interruption of the pathways for synthesizing by-products. The implementation of the above-mentioned strategies involves a wide variety of DNA manipulations, including site-directed mutagenesis (SDM), gene inactivation and over-expression. In contrast to conventional breeding via repeated physical and/or chemical mutagenesis, genetic engineering breeding gives more practicable options for subsequently isolating mutant strains because it can avoid poor physiological characteristics and unknown inherited characteristics. Therefore, developing a new method to achieve faultless DNA manipulations is one of the most popular research subjects.
Here we review the common strategies used for DNA manipulations. We focus on the strategies for DNA manipulations via genetically modifying the E. coli and C. glutamicum genomes. In addition, the potential problems of multi-layered DNA manipulations are considered. However, this review avoids any discussion of plasmid-mediated gene over-expression. It intends to be an easy introduction for novices and a source of new experimental information for experienced investigators.
2 Strategies for SDM
SDM, also called “rational mutagenesis” (Mandaci, 2011), is commonly used to introduce mutations at definite sites of a target DNA fragment, including the genome and plasmid, via polymerase chain reaction (PCR) or restriction endonuclease reaction (RER). It plays a great role in understanding the regulatory motifs of operon and the relationship of protein structure to function (Ling and Robinson, 1997; Seyfang and Jin, 2004; Wu et al., 2013). Depending on the numbers of mutational sites, SDM can be divided into two types: single site-directed mutagenesis (Single-SDM) and multiple site-directed mutagenesis (Multi-SDM) (Liang et al., 2012).
2.1 Strategies of Single-SDM
Single-SDM is mainly based on the amplification of double-stranded DNA (dsDNA) plasmid using complementary primer pairs, which contain 20–30 oligonucleotides and the desired mutation (Ling and Robinson, 1997; Holland et al., 2015). Because of its simplicity, time-saving capability, and relatively high efficiency, this approach has become a common strategy for introducing mutation into E. coli and C. glutamicum genomes (Ling and Robinson, 1997; Muyrers et al., 2001; Xu et al., 2014b). Although most of the methods used for Single-SDM have been reviewed by researchers around 15–20 years ago (Chatellier et al., 1995; Ling and Robinson, 1997; Muyrers et al., 2001), many new methods are springing up along with the development of genetic engineering technique.
2.1.1 Single-SDM by enzyme digestion and homologous recombination (ED-HR)
Commercial kits are simple and easy to use for SDM (Imai et al., 1991; Martin et al., 1995; Chiu et al., 2004; Stoynova et al., 2004), but some defects usually limit their application in creating larger deletions (Li et al., 2008). In order to overcome the limitations of commercial kits, the methods used for larger deletion have been developed based on DNA ligation and hybridization in vitro (Imai et al., 1991; Chiu et al., 2004; Stoynova et al., 2004) or on homologous recombination in vivo (Martin et al., 1995). The improved homologous recombination technique has greatly simplified the procedure of mutagenesis (Li et al., 2008), with only two steps: PCR amplification and transformation (Martin et al., 1995). However, the efficiency is not invariable because the plasmid used as a template in PCR amplification is also transformed into a competent cell (Li et al., 2008). Although many efforts have been made to overcome this obstacle, such as gel purification of PCR products, they are time-consuming and do not work well for purifying the few products resulting from fewer PCR cycles and/or lower amplification efficiency (Li et al., 2008).
It is generally known that most bacteria possess DNA restriction-modification (R-M) systems that are used to protect the cell from intrusion by foreign DNA. In most cases, “self” DNA is marked by methylation, whereas in “non-self” DNA it is absence of methylation which marks it (Siwek et al., 2012). The DpnI restriction system is one of the R-M systems (Johnston et al., 2013). DpnI endonuclease cleaves only the double-stranded (ds)-methylated 5′-GmATC-3′ sequence in DNA (Lacks and Greenberg, 1977). Interestingly, the plasmid extracted from bacteria contains methyladenosine at the DpnI restriction site, which makes the plasmid susceptible to DpnI (Lu et al., 2002), whereas PCR-amplified DNA does not contain methyladenosine (Li et al., 2008). Based on this principle, PCR using plasmid DNA as a template is a very useful way to obtain replication and mutagenesis (Lu et al., 2002). PCR products digested by DpnI have been widely used in mutagenesis, and the protocols are listed in Fig. 1a. In general, the PCR products are digested in vitro by DpnI (Weiner et al., 1994; Martin et al., 1995; Qi and Scholthof, 2008). Li et al. (2008) pointed out a simple and rapid strategy to digest PCR products in vivo by DpnI when carrying out SDM. The crucial aspects of this strategy are inactivation of dam gene in chromosome and heterologous expression of dpnC gene (encoding DpnI) in the host cell. Since DpnI was expressed in vivo, there is no need to purchase DpnI and to manipulate PCR products.
Schematic diagrams of the Single-SDM method for base deletion, substitution, and insertion by enzyme digestion and homologous recombination (a) (Li et al., 2008) and by enzyme ligation and homologous recombination (b) (Wu et al., 2013)
Blue lines represent targeted gene; bright-green lines represent the homologous fragment between target gene and primers; red lines represent desired mutant sites; pink lines represent restrict enzyme sites (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
2.1.2 Single-SDM by enzyme ligation and homologous recombination (EL-HR)
The above-mentioned ED-HR and other PCR-based traditional Single-SDMs (Picard et al., 1994; Brøns-Poulsen et al., 1998; Chapnik et al., 2008; Li et al., 2008) require at least two PCR rounds, multiple steps of enzymatic treatment, DNA template purification, and primer transformation or purification via high-performance liquid chromatography (HPLC) (Picard et al., 1994; Qi and Scholthof, 2008; Wan et al., 2012). In addition, the targeted gene should be sub-cloned into an original plasmid to form targeted plasmid which is used as a template to amplify the mutant plasmid with the desired mutation site (Picard et al., 1994). However, undesired amplification of these methods results in long extension time and low mutagenesis efficiency (Wang et al., 2011; Adachi and Fukuhara, 2012). Moreover, these methods require the digestion of the PCR products with DpnI in vitro or in vivo (Chapnik et al., 2008; Li et al., 2008; Qi and Scholthof, 2008). All these can be expensive and time-consuming (Wan et al., 2012).
Many plasmids, including T-vector (e.g. pUC18 and pMD™18-T), mobilizable plasmid (e.g. pK18mobsacB and pK19mobsacB), and expression plasmid (e.g. pDXW-8 and pET28a), contain a multiple cloning site (MCS) (Schäfer et al., 1994; Xu et al., 2010). The MCS can be used to clone the targeted DNA fragment into these plasmids. Based on the circularization of PCR products via homologous recombination in vivo (Jones and Winistorfer, 1991), Wu et al. (2013) proposed a rapid and efficient protocol depending on two separate PCR amplifications and one-step ligation, which we call an “EL-HR”. This protocol can be used to introduce deletions, insertions, and substitutions into any site of the target genes (Fig. 1b). Since the up-fragment and down-fragment (i.e. A fragment and B fragment) of the targeted gene are independently amplified with the same PCR parameters and at the same time, it avoids long extension time, high PCR error rate, and low mutagenesis efficiency. In addition, this protocol does not need the digestion of the parental template because the targeted plasmid is constructed by enzyme ligation rather than by PCR.
2.1.3 Single-SDM by TA strategy
Although these PCR-based approaches are useful and powerful for SDM, there are still many defects: (1) long or HPLC-purified primer (Davis et al., 1999; Salerno et al., 2005); (2) high-fidelity thermostable DNA polymerase and restriction endonuclease (Salerno et al., 2005); (3) the need for subcloning the desired gene into the original plasmid to form a targeted plasmid (Davis et al., 1999); (4) undesired amplification of the wild-type gene template (Ke and Madison, 1997); (5) the self-annealing of megaprimer strands (Brøns-Poulsen et al., 1998; Siloto and Weselake, 2012); (6) the unbalanced melting temperatures of the primers (Tseng et al., 2008; Saeedi et al., 2012); (7) the requirement for a high concentration of megaprimer to ensure the successful modification (Wan et al., 2012); (8) the requirement for additional steps for preparation of the template plasmid (Urban et al., 1997) or restriction endonuclease treatments of PCR products (Seraphin and Kandels-Lewis, 1996).
Alternatively, Adachi and Fukuhara (2012) proposed a simple method using TA strategy with synthetic mutagenic oligonucleotides. By application of this strategy, virtually any DNA manipulation can be achieved, and it is even possible to introduce foreign sequences into the PCR-amplified 3′ A-overhang vector in a size-independent and site-specific manner (Adachi and Fukuhara, 2012). The protocols of this method are illustrated in Fig. 2 and the crucial aspects are as follows:
-
(1)
The choice of thermostable DNA polymerases. 3′→5′ exonuclease exists inherently in DNA polymerase γ and T4 polymerase (Khare and Eckert, 2002), and involves rectification of a mismatched base-pair. Therefore, in order to leave 3′ A-overhang after PCR, we should select 3′→5′ exonuclease-deficient thermostable DNA polymerases for PCR.
-
(2)
The synthetic mutagenic ds-oligonucleotides. According to the principle of adenine-thymine (AT) base pairs, the 3′ A-overhang vector and 3′ T-overhang synthetic mutagenic ds-oligonucleotides are connected. Construction of a different mutation type is executed via synthesizing the different mutation type ds-oligonucleotides with the 3′ T-overhang sticky ends.
-
(3)
The length of the targeted plasmid with target gene. PCR-mediated amplification increases the risk of introducing undesired mutation during use of thermostable polymerase, especially use of low-fidelity thermostable polymerase (Wu et al., 2013). Moreover, Adachi and Fukuhara (2012) have pointed out that excessively long extension time increases the likelihood of error rates and of generating artifacts.
-
(4)
The storage time of PCR product. Because 3′ A-overhangs on the PCR product would be degraded over time to reduce the ligation efficiency with 3′ T-overhang synthetic mutagenic ds-oligonucleotides, the fresh PCR product should be used if the SDM has been done.
Schematic diagram of the TA strategy-based Signle-SDM method for base deletion, substitution, and insertion (Adachi and Fukuhara, 2012)
Blue lines represent targeted gene; red lines represent mutant sites; 5′→3′ represents base direction (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
2.1.4 Comparison of major methods for Single-SDM
With the development of genetic engineering technology, numerous novel Single-SDM methods based on the QuikChange™ SDM system (QCM)-like kits or previous protocols have been developed to overcome the disadvantages of previous methods (Ke and Madison, 1997; Urban et al., 1997; Salerno et al., 2005; Qi and Scholthof, 2008; Tseng et al., 2008; Wang et al., 2011; Adachi and Fukuhara, 2012; Wan et al., 2012; Sun et al., 2015). Most of these methods can be widely used to construct various difficult-to-construct mutants in areas where traditional SDM protocol cannot perform. Moreover, these methods can vastly simplify routine work and reduce experimental cost. However, there are no perfect methods for Single-SDM, including the above-mentioned methods, as far as we know. In summary of the sections on the applications of PCR-based approaches and non PCR-based approaches in Single-SDM, some aspects of above-mentioned methods are compared in Table 1.
2.2 Strategies of Multi-SDM
As stated previously, SDM plays an important role in genetic research, such as understanding the regulation site of operon or the relationship of protein structure to function (Ling and Robinson, 1997; Seyfang and Jin, 2004; Wan et al., 2012). Therefore, many strategies used for Single-SDM are springing up with the development of genetic engineering technology (see Section 2.1). However, it is sometimes necessary to modify several sites in one amino acid sequence to understand the structure and function of a given protein. Hence, a rapid and efficient Multi-SDM strategy would be of great benefit in various applications in functional proteomics and genetic engineering, such as codon optimization for heterologous expression, generation of cysteine-less proteins for subsequent cysteine-scanning mutagenesis, and disulfide-scanning mutagenesis studies (Davis and Seqaloff, 2002; Wang et al., 2012) or (re)design and removal of restriction endonuclease sites in expression vectors (Seyfang and Jin, 2004). Many strategies have been developed to simplify the generation of multiple mutations in a targeted gene.
2.2.1 Multi-SDM by multiple mutagenic oligonucleotide-directed (MMOD) method
Oligonucleotide-directed SDM methods have been described, including PCR-based methods, and methods based on plasmid DNA templates and mutant DNA (Hogrefe et al., 2002). Most of these methods are used to introduce only one mutation site at a time. In 1990, Perlak (1990) presented evidence that it is possible to modify multiple sites in a targeted DNA by the oligonucleotide-directed SDM method using multiple mutagenic oligonucleotides as primers. We call this a “Multiple Mutagenesis Oligonucleotide-Directed (MMOD) method”. The theoretical basis of this method is that the mutant strand synthesized by primer extension can be mediated by T4 DNA polymerase or by a Klenow fragment of DNA polymerase I. T4 DNA polymerase cannot remove the mutagenic primer, whereas the Klenow fragment can be used to displace the primer oligonucleotide which contains the desired mutations and homologous fragment of the original strand (Nossal, 1974). After a series of PCR reactions with multiple mutagenic oligonucleotides as primers, the mutants with desired mutations are screened. Tu and Sun (1996) have successfully introduced three mutation sites in a targeted gene. However, the efficiency of mutagenesis is low (Perlak, 1990).
2.2.2 Multi-SDM by adaptation methods based on QuikChange™
The QuikChange™ protocol, developed by Stratagene (La Jolla, CA, USA), is one of the simplest and fastest methods for introducing mutations (Hogrefe et al., 2002). QuikChange™ protocol uses a reverse complement mutational primer pair to replicate a template plasmid with the targeted gene, and then introduces mutation(s) at the binding site(s) for primer. It relies on the fact that DNA synthesized in vitro is not methylated (Li et al., 2008), whereas the template plasmid extracted from E. coli cell is methylated (Lu et al., 2002), and the methylated DNA can be digested by DpnI. However, there are several deficiencies (Mitchell et al., 2013): (1) DNA replication is linear rather than exponential; (2) it needs high-fidelity DNA polymerase; (3) it is hard to introduce multiple mutations in a single reaction; (4) efficiency is limited by the size of the template plasmid; (5) it has high error rates due to long extension time (Adachi and Fukuhara, 2012); (6) DpnI is required to digest the template DNA; (7) primers are synthesized with a 5′ phosphate moiety and purified by polyacrylamide gel electrophoresis (PAGE) (Hogrefe et al., 2002); (8) the self-annealing and self-pairing need to be carried out during primer design (Liu and Naismith, 2008); (9) it is hard to carry out deletions and insertions. In order to address these shortcomings, many researchers have proposed new strategies.
Kirsch and Joly (1998) divided the in vitro DNA synthesis into two successive phases, and the two newly amplified DNA fragments served as “megaprimers” to synthesize the remainder of plasmid. This method can be used with non-overlapping oligonucleotides and non-specific restriction sites, and achieves simultaneously two separate mutation sites in a gene. By the use of in vitro dam-methylation among the successive PCR amplifications, Kim and Maas (2000) pointed out that all mutations would be introduced after a single transformation and DNA preparation step. This adaptation method not only saves time but also preserves high targeting efficiency. In addition, this method plays an important role when intermittent phenotypic selection is unfavorable, or the targeted gene is unstable or tends to recombine in vivo. Moreover, Liu and Naismith (2008) used novel primers to achieve multiple mutations in a gene. The primers, containing extended non-overlapping sequences at the 3′ end and primer-primer complementary sequences at the 5′ end, promote primer-template annealing via eliminating primer dimerization and permit the newly-synthesized DNA to be used as the template in subsequent PCR cycles.
Multichange isothermal (MISO) mutagenesis is another new protocol to introduce Multi-SDM into plasmid DNA, which is based on the QuikChange™ protocol and one-step isothermal (ISO) assembly (Mitchell et al., 2013). This method overcomes all of the aforementioned defects of the traditional QuikChange™ protocol. Fig. 3 shows an overview of this protocol. The MISO mutagenesis can be used as a simple strategy for making three types of DNA manipulations (i.e. base substitution, insertion, and deletion) in a single round of experimentation (Adachi and Fukuhara, 2012). However, there are still several limitations, such as DNA sequence, the error rate associated with oligonucleotide synthesis, and mis-assembly errors from the ISO-assembly step. In addition, it is difficult to introduce mutations at nearby bases.
Schematic diagram of the multichange isothermal mutagenesis for Multi-SDM (a) and one-step isothermal assembly relies on the concerted action of three enzymes (b) (Mitchell et al., 2013)
Blue lines represent targeted gene; red lines represent insertion sequences; the same colour lines represent homologous regions; symbols “^” represent mutant sites; orange box represents deletion region (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
2.2.3 Multi-SDM by overlap extension PCR (OE-PCR)
OE-PCR has been widely used to introduce a single mutation embedded in the oligonucleotide primers (Ho et al., 1989) and to change gene (Horton et al., 1990). Based on the principle and strategy of OE-PCR in Single-SDM, OE-PCR has been developed for use in Multi-SDM (Urban et al., 1997; Tian et al., 2010; Luo et al., 2012; Wäneskog and Bjerling, 2014). In traditional OE-PCR, two or more mutation-embedding fragments are separately amplified using internal primers which contain mutation sites and an overlapping sequence. These fragments are fused together and then form a new double strand template DNA in a subsequent extension reaction, in which the 3′ overlap of each strand serves as primer for extending the 3′-end of the complementary strand. Finally, the resulting mutant DNA is generated from the fused product by PCR using external oligonucleotide primers (Luo et al., 2012). Compared with the Quick-change method, traditional OE-PCR is good for Multi-SDM with respect to the linear template. However, it is necessary to remove the wild-type DNA template and residual primers in the first amplification (Peng et al., 2006). This purification step is time-consuming and expensive. In order to avoid the purification step, Tian et al. (2010) have developed a method which is based on OE-PCR and DpnI digestion. In this method, the circular plasmid with a targeted gene rather than linear DNA was used as template. The key point of this method is that the template plasmid is required to be methylated by Dam methyltransferase or extracted from dam+ E. coli (Tian et al., 2010). Despite efforts to continuously improve the application of OE-PCR, there are still some defects, such as it being hard to manipulate large DNA fragments (i.e. >7 kb).
2.2.4 Multi-SDM by multiple rounds PCR (MR-PCR)
PCR-based mutagenesis protocols have been widely used to change an amino acid from one to another in recent years. Several protocols have been developed to achieve the desired mutation using PCR technology (Ling and Robinson, 1997). The mutation introduced into the gene is executed by MR-PCR with multiple mutagenic oligonucleotides as primers (Meetei and Rao, 1998). In contrast to the aforementioned three methods, this method does not require DNA ligase or 5′ phosphorylation of mutagenic primers, which is concerned with annealing efficiency of the mutagenic primer and 5′ to 3′ exonuclease activity of thermostable DNA polymerase, and it needs to subclone the PCR product each time (Meetei and Rao, 1998). One of the key points of this method is to have two templates, i.e. “A” and “B”. “A” has an extra flanking sequence except for the gene or part of the targeted gene, which is used as a binding site for one of the flanking primers (S1), whereas “B” does not have this sequence. “B” either has a different flanking fragment or lacks the flanking sequence with few mismatches at the end of the flanking fragment (Barettino et al., 1994; Boles and Miosga, 1995). It should be noted that the template “A” must be removed by gel electrophoresis after the first round of PCR.
2.2.5 Multi-SDM by primer extension and ligation (PEL)
As described previously, multiple mutagenic oligonucleotides are used as primers to execute Multi-SDM by MR-PCR (Tu and Sun, 1996; Patel et al., 2009). However, the efficiency of mutagenesis is lower (Patel et al., 2009). Moreover, these methods require two pairs of complementary primers to introduce one mutation (Seyfang and Jin, 2004). To avoid the use of two complementary primers and to overcome the shortcomings of MR-PCR, Seyfang and Jin (2004) developed a simple and rapid method for SDM, which can be used to achieve more than 10 mutations with up to 100% efficiency. The fragment with the mutation is synthesized by PEL with T4 DNA polymerase and ligase, respectively. Then the fragment with the mutation is amplified by high-fidelity PCR with specific tailed anchor primers which contain a unique 25-nucleotide tail for subsequent mutational-fragment-specific PCR and three restriction endonuclease sites for subcloning the PCR product into different vectors. The overview of this method is given in the report of Seyfang and Jin (2004). In contrast to the above-mentioned methods, this method does not require the synthesis of single-stranded DNA as template and benefits from the use of only one antisense mutagenic primer for introducing one mutation (Seyfang and Jin, 2004). And it only needs a single round of PCR to introduce all of the mutation.
2.2.6 Multi-SDM by amplification, ligation, and suppression PCR (MALS)
In contrast to other PCR-based strategies for multi-SDM, Fushan and Drayna (2009) developed a rapid and efficient method for the introduction of a wide variety of mutations. The generation of mutation types is dependent on the use of the designed internal oligonucleotides, and the desired mutation exists at the 5′ end of one or both internal oligonucleotides (Fushan and Drayna, 2009). An outline of this MALS-mediated strategy is shown in Fig. 4a. The novelty of this method is that the mutation is introduced using SDM in combination with suppression PCR. The procedure consists of sequential rounds, and each individual round requires PCR amplification of the target DNA with two pairs of non-overlapping primers (Fushan and Drayna, 2009). Three types of molecules, i.e. homomeric ligation products, non-ligated molecules, and heteromeric ligation products, are found in the ligation products (Fig. 4b). The effect of suppression PCR in preventing PCR amplification (Diatchenko et al., 1996; Dai et al., 2007; Liew et al., 2015), is that the suppression PCR is used to screen the desired mutant molecules from other types of molecules. It is important to note that the mutagenic primers must be phosphorylated at the 5′ end.
Outline of this MALS-mediated strategy and three types of molecules after mixing and ligation of two PCR products with SO1/IR1 and IF1/SO2 as primers, respectively (Fushan and Drayna, 2009)
Black lines represent targeted gene; bright-green and blue lines represent homologous arms; red lines represent mutant sites; Ⓟ represents phosphate groups (Note: for interpretation of the references to color in this figure legend, the reader is referred to the web version of this article)
2.3 Strategies to design SDM primers
The efficiency of the oligonucleotide-mediated SDM is greatly affected by the design of mutagenic oligonucleotide primers and the quality of the subsequent primer-template annealing. At present, the design of primers is based on an optimized strategy using partial- or complete-overlapping primers (Zheng et al., 2004; Xu et al., 2013). Therefore, the design of SDM primers has become even more important in SDM, and especially in Multi-SDM (Seyfang and Jin, 2004). In order to distinguish the mutant sequence from the wild-type gene, the main strategy is to introduce new or remove previous restriction sites and then add a unique identifier to each mutation during the design of the SDM primers. However, it is hard to distinguish the different point mutations of the same wild-type gene (Karnik et al., 2013). Therefore, a unique restriction site (also known as a “silent” restriction site) specified for each mutation should be used as a simple and reliable identifier. There are four primer design programs that allow the introduction of a “silent” restriction site during SDM primer design, i.e. SILMUT (Shankarappa and Vijayananda, 1992), Primer Generator (Turchin and Lawler, 1999), Site-Find (Evans and Liu, 2005), and SDM-Assist (Karnik et al., 2013). In contrast to Primer Generator and SiteFind, SDM-Assist overcomes the limitation of Primer Generator which only runs in a DOS-based text-entry intensive interface (Turchin and Lawler, 1999), and the defects of SiteFind which only allows the input of about 400 bp nucleotides of DNA (Evans and Liu, 2005). Moreover, SDM-Assist also can be used to manipulate the input sequence in a user-friendly format and to help the design of the primers and the calculation of their thermodynamic parameters (Karnik et al., 2013).
3 Strategies for gene inactivation
SDM can be used for deletion of bases, but this mutation is a small deletion, no more than 170 bp (Qi and Scholthof, 2008; Wan et al., 2012; Wu et al., 2013). That is in stark contrast to gene knockout which is used to completely or partly delete one gene to inactivate the function of this gene, up to 50 kb (Sawitzke et al., 2013). Gene inactivation plays an important role in revealing the function of specific genes and in developing novel variants (Wu et al., 2013). With its sequence interrupted, the target gene would be translated into a nonfunctional protein that leads to phenotypic modulation in most cases. This strategy is very useful for classical genetic research and for modern biotechniques including functional genomics (Sawitzke et al., 2013). In general, gene inactivation is based on the theories of homologous recombination to replace the existing autologous gene with a designed heterogenous gene. With the development of gene inactivation, new techniques are gradually being introduced in gene inactivation, such as insertional inactivation.
3.1 Gene inactivation based on homologous recombination
Homologous recombination-mediated gene inactivation is an in vivo genetic modification. In contrast to in vitro genetic modification, it is not limited by the location of the enzyme digestion sites and the construction of base-pairs (Sawitzke et al., 2013). It is executed by introducing recombinant DNA (plasmid or linear DNA) containing selectable marker(s), counter-selectable marker(s), and homologous arm(s) into cells (Datsenko and Wanner, 2000; Yu and Ellis, 2000; Muyrers et al., 2001; Nakashima and Miyazaki, 2014). Recombinant DNA can be constructed in vitro and then introduced into cells. Homologous recombination occurs through homologous arm(s), and the recombination strains are selected according to the selectable marker(s) and counter-selectable marker(s).
3.1.1 Plasmid-mediated homologous recombination
Research on plasmid-mediated homologous recombination showed some successful examples using intermolecular recombination between two circular molecules (Poustka et al., 1984; Philippe et al., 2004; Xu et al., 2013; 2014b; 2015). In this method, the host cell must have the genetic background of recA+, which is called RecA-dependent strategy. An outline is shown in Figs. 5a and 5b. Before the first round of homologous recombination, a dedicated plasmid with several components must be constructed. These components include: (1) selectable marker(s) and counter-selectable marker(s); (2) long homologous arm(s); and, (3) controlling element(s). To avoid selectable markers or scars in the chromosome, the most useful tools are suicide plasmids (Philippe et al., 2004). There are three types of suicide plasmids that have been used to construct recombinant E. coli or C. glutamicum strains: temperature-sensitive plasmids, e.g. pSC101 and its derivatives (Cornet et al., 1994), and plasmids carrying the replication origin of R6K, such as pCVD441 and its derivatives (Donnenberg and Kaper, 1991), or the replication origin of pMB1, such as pK18mobsacB and its derivatives (Schäfer et al., 1994). These systems involve a two-step procedure: recombinant targeted plasmid integrates in the targeted sequence by the first round of homologous recombination, then excises from the chromosome via the second round. Excision of the recombinant plasmid from the chromosome is executed with counter-selectable markers, because the cell would die in the presence of a counter-selectable compound if the plasmid remains in the chromosome (Reyrat et al., 1998). There are three major counter-selectable markers: (1) fusaric acid-sensitivity system (Maloy and Nunn, 1981); (2) streptomycin-sensitivity system (Dean, 1981); (3) sucrose-sensitivity system (Gay et al., 1983).
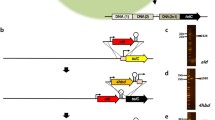
Schematic diagram of gene inactivation based on plasmid-mediated homologous recombination (a, b) and linear DNA-mediated homologous recombination (c, d)
sm represents selectable marker; csm represents counter-selectable marker; ce represents controlling element; A represents the autologous A gene; A-L represents the left homologous arm of A gene; A-R represents the right homologous arm of A gene; B represents the extraneous B gene; C represents the C gene; P1 and P2 represent the primers according the sequence of antibiotic marker
Much research has reported that E. coli could be genetically modified to produce various amino acids and organic acids by plasmid-mediated homologous recombination (Zhou et al., 2003; Causey et al., 2004; Imaizumi et al., 2005; Zhang et al., 2007). At present, as far as we are aware, gene inactivation of the C. glutamicum genome is mainly carried out by plasmid-mediated homologous recombination. Over the past decade, many researchers dedicated themselves to develop the new methods of plasmid-mediated homologous recombination to overcome the defects in the present method (Poustka et al., 1984; Homilton et al., 1989; Kato et al., 1998; Philippe et al., 2004). However, there remain questions on the requirement of long homologous arm(s), how time-consuming it is, low success rates, and the limitation of the recombinogenic window (Muyrers et al., 2001).
3.1.2 Linear DNA-mediated homologous recombination
An alternative homologous recombination strategy was developed where the recombination is performed by introducing recombinant linear DNA rather than a plasmid. In addition, this strategy is not dependent on RecA, but it is mediated by phageencoded recombinant proteins, either RecE/RecT from Rac phage (Zhang et al., 1998) or Redβ (encoding Bet), Redexo (encoding Exo), and Redγ (encoding Gam) from λ phage (Datsenko and Wanner, 2000; Yu and Ellis, 2000). RecE and Exo are 5′→3′ dsDNA specific exonucleases, and are required for dsDNA recombination. RecT and Beta are single-stranded DNA (ssDNA) annealing proteins, and are the central recombinases in homologous recombination. Gam is not absolutely required for recombination but promotes dsDNA recombination because it inhibits the host RecBCD exonuclease that is normally used to digest linear dsDNA (Sawitzke et al., 2013).
An outline of this linear DNA-mediated strategy is shown in Figs. 5c and 5d. This strategy possesses the advantages inherent in the use of linear targeted DNA and thus increases the success rate. A linear targeted DNA carries two short homologous arms (35–60 bp) flanking a selectable gene. The shorter homologous arms can be made by oligonucleotide synthesis rather than PCR and thus offers a shorter operation time. In addition, the linear targeted DNA is obtained by PCR and no dedicated plasmid needs to be constructed (Muyrers et al., 2001). In particular, PCR products carry selectable and counter-selectable gene(s) that can be used to select recombinants without a marker gene. However, unintended additional homologous recombination may occur in the targeted DNA that limits the recombinogenic window. In addition, residual scar, such as flippase (FLP) recognition target (FRT), would disturb the next chromosomal rearrangement in FLP-promoted recombination events (Datsenko and Wanner, 2000).
3.2 Gene inactivation based on insertional inactivation
Insertional inactivation has been used for inactivating the prokaryotic (such as E. coli and C. glutamicum) and eukaryotic (such as yeast) genes for many years. Several methods have been reported that can achieve the desired gene-inactivation mutants by inserting foreign sequences into a target gene. The first method is based on allelic exchange. The targeted fragment with a selectable marker is constructed in vitro and then transferred into the host cell. The artificial fragment replaces the targeted wild-type gene by double-crossover homologous recombination (Tilly et al., 2000; Xu et al., 2014c). The key aspect of this method is that it requires a gene transfer mechanism and a selectable marker (Tilly et al., 2000). The second method is carried out by plasmid integration. The plasmid for integration must contain a selectable marker and a part of the targeted gene, which could result in reversal of the integration by a single-crossover recombination. However, it is hard to maintain the selection for the plasmid, and plasmid integration can also occur without mutation at the insertion site (Tilly et al., 2000).
In contrast to being dependent on homologous recombination, the third method used for gene inactivation is random insertion in the genome using a transposable element (Berg and Berg, 1996). This method can easily generate large-scale disruption of the genome (Judson and Mekalanos, 2000; Suzuki et al., 2006). Transposons are mobile genetic elements that can transpose from one location in a genome to another (Judson and Mekalanos, 2000). Transposon insertions can alter the regulation and expression of genes, so the application of it has provided valuable functional genomic information to complement genome-sequencing projects (Kim, 2015). However, the drawbacks of this method are that it is difficult to identify the transposon insertion sites, and requires complex procedures (Suzuki et al., 2006). In order to overcome these drawbacks, Suzuki et al. (2006) have developed a method which can be used to simply identify the transposon insertion sites based on the sequences of thermal asymmetric interlaced (TAIL)-PCR products of mutant cells by using BLAST and Per1.
All in all, the three above methods of insertional inactivation require a usable selectable marker. The outline of these methods is similar to that of homologous recombination-mediated gene inactivation, only with a selectable marker in the genome.
4 Strategies for gene over-expression
Plasmid-mediated structural gene over-expression is a common strategy in genetic modification of E. coli and C. glutamicum (Ikeda and Katsumata, 1998). Plasmids generally carry genetic markers, such as a drug resistance marker, which are used for high-efficiency screening of target-transformants (Xu et al., 2014c). However, there are several disadvantages to the application of plasmid-mediated gene over-expression, such as the instability of the plasmid and negative effect on cell growth. Although Hu et al. (2014) have constructed a novel expression system for gene amplification using plasmid, the antibiotic resistance gene is also introduced into the cell. In past decades, constructions of integrative plasmids which carry a homologous chromosomal fragment and drug resistance marker have made it possible to insert a gene into the chromosome. The homologous fragment in a plasmid consists of multiple copies in the chromosome, such as IS13869 (Amador et al., 2000) and rRNA gene (Correia et al., 1996). The integration is carried out by single-crossover recombination between the integrative plasmid and the recipient genome occurring in the homologous region (Ikeda and Katsumata, 1998). In addition, Suzuki et al. (2006) have reported a new Cre/mutant lox system for integrating the heterologous genes into the C. glutamicum genome. However, the host cell must contain a mutant lox gene, and the integrated foreign gene must contain a selective marker, such as antibiotic resistance genes (Suzuki et al., 2006). Recently we have developed a method for the simultaneous replacement of a targeted gene by a given gene cassette, leaving no genetic markers (Fig. 6) (Xu et al., 2014c). However, the methods mentioned above are only used in modifying the C. glutamicum chromosome. Although gene replacement is one of the easiest procedures for gene manipulation in E. coli, there is no method for gene over-expression by inserting into the E. coli chromosome. It is a possible reason why it is hard to screen the target transformants.
Strategy used for genetic manipulation in Corynebacterium glutamicum (Xu et al., 2014c)
A represents the autologous A gene; A-R represents the right arm of A gene; A-L represents the left arm of A gene; Ptac-B-rrnBT1T2 represents insertional cassette; Ptac represents the promoter tac; B represents the extraneous B gene; rrnBT1T2 represents the terminator rrnBT1T2; kan represents kanamycin resistance encoding gene; sacB represents levansucrase encoding gene
5 Conclusions and perspectives
Over the last several years, genetic engineering techniques have been widely employed in research and development for strain improvement. Genetic engineering is the name of a group of techniques used for direct genetic modification of organisms, such as SDM, gene inactivation, and gene over-expression (Montaldo, 2006). The strategies presented outline how precise DNA modifications, including SDM, gene inactivation, and gene over-expression, can be made to DNA molecules in E. coli and C. glutamcum of any size. This review sums up the latest methods used for genetically modifying E. coli and C. glutamicum genome, and discusses the technical problems for multi-layered DNA manipulations. In particular, key factors have been described for each method to perform the DNA manipulation. From this review, we can see many efficient methods for genetically modifying E. coli and C. glutamicum genome. Choices greatly depend on the experimental needs and the resources of the researchers.
It should be noted that, as far as we are aware, the methods used for gene over-expression in the genome are very rare, especially for E. coli. Plasmid-mediated structural gene over-expression is a common strategy in the genetic modification of E. coli and C. glutamicum (Ikeda and Katsumata, 1998). However, plasmid-mediated gene over-expression will inevitably introduce a drug resistance marker, which increases the concern over the distribution of antibiotic resistant genes (Tauch et al., 2002). Considering the very broad interest generated by this subject, we can reasonably hope that strategies will be found to counteract this deficiency, and that new approaches will be developed. At the same time, it is crucial to construct stable high-yielding strains.
Compliance with ethics guidelines
Jian-zhong XU and Wei-guo ZHANG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Amador, E., Franciso, J., Castro, J.M., 2000. A Brevibacterium lactofermentum 16S rRNA gene used as target site for homologous recombination. FEMS Microbiol. Lett., 185(2):199–204. http://dx.doi.org/10.1111/j.1574-6968.2000.tb09062.x
Adachi, Y., Fukuhara, C., 2012. TA strategy for rapid and efficient site-directed mutagenesis. Anal. Biochem., 431(1): 66–68. http://dx.doi.org/10.1016/j.ab.2012.08.030
Baba, T., Ara, T., Hasegawa, M., et al., 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol., 2:2006–2008. http://dx.doi.org/10.1038/msb4100050
Berg, C.M., Berg, D.E., 1996. Transposable element tools for microbial genetics. In: Neidhardt, F.C., Curtiss III, R.C., Ingraham, J.L., et al. (Eds.), Escherichia coli and Salmonella. ASM Press, Washington, DC,USA, p.2588–2612.
Barettino, D., Feigenbutz, M., Valcárcel, R., et al., 1994. Improved method for PCR-mediated site-directed mutagenesis. Nucleic Acids Res., 22(3):541–542. http://dx.doi.org/10.1093/nar/22.3.541
Boles, E., Miosga, T., 1995. A rapid and highly efficient method for PCR-based site-directed mutagenesis using only one new primer. Curr. Genet., 28(2):197–198. http://dx.doi.org/10.1089/pho.2008.2449
Brøns-Poulsen, J., Petersen, N., Horder, M., et al., 1998. An improved PCR based method for site directed mutagenesis using megaprimers. Mol. Cell. Probe., 12(6):345–348. http://dx.doi.org/10.1006/mcpr.1998.0187
Becker, J., Zelder, O., Häfner, S., et al., 2011. From zero to hero—design-based systems metabolic engineering of Corynebacterium glutamicum for L-lysine production. Metab. Eng., 13(2):159–168. http://dx.doi.org/10.1016/j.ymben.2011.01.003
Causey, T.B., Shanmugam, K.T., Yomano, L.P., et al., 2004. Engineering Escherichia coli for efficient conversion of glucose to pyruvate. PNA., 101(8):2235–2240. http://dx.doi.org/10.1073/pnas.0308171100
Chapnik, N., Sherman, H., Frog, O., 2008. A one-tube sitedirected mutagenesis method using PCR and primer extension. Anal. Biochem., 372(2):255–257. http://dx.doi.org/10.1016/j.ab.2007.07.020
Chatellier, J., Mazza, A., Brousseau, R., et al., 1995. Codonbased combinatorial alanine scanning site-directed mutagenesis: design, implementation, and polymerase chain reaction screening. Anal. Biochem., 229(2):282–290. http://dx.doi.org/10.1006/abio.1995.1414
Chiu, J., March, P.E., Lee, R., et al., 2004. Site-directed, ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res., 32(21):e174. http://dx.doi.org/10.1093/nar/gnh172
Cornet, F., Mortier, I., Patte, J., et al., 1994. Plasmid pSC101 harbors a recombination site, psi, which is able to resolve plasmid multimers and to substitute for the analogous chromosomal Escherichia coli site dif. J. Bacteriol., 176(11):3188–3195.
Correia, A., Martin, J.F., Castro, J.M., 1996. Targeted integration of foreign genes into repetitive sequences of the Brevibacterium lactofermentum chromosome. FEMS Microbiol. Lett., 142(2–3):259–264. http://dx.doi.org/10.1111/j.1574-6968.1996.tb08440.x
Dai, Z.M., Zhu, X.J., Chen, Q., et al., 2007. PCR-suppression effect: kinetic analysis and application to representative or long molecule biased PCR-based amplification of complex samples. J. Biotechnol., 128(3):435–443. http://dx.doi.org/10.1016/j.jbiotec.2006.10.018
Datsenko, K.A., Wanner, B.L., 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. PNA., 97(12):6640–6645. http://dx.doi.org/10.1073/pnas.120163297
Davis, D.P., Seqaloff, D.L., 2002. N-linked carbohydrates on G protein-coupled receptors: mapping sites of attachment and determining functional roles. Methods Enzymol., 343:137–156. http://dx.doi.org/10.1016/S0076-6879(02)43136-9
Davis, M.D., Wonderling, R.S., Walker, S.C., et al., 1999. Analysis of the effects of charge cluster mutations in adeno-associated virus Rep68 protein in vitro. J. Virol., 73(3):2084–2093.
Dean, D., 1981. A plasmid cloning vector for the direct selection of strains carrying recombinant plasmids. Gen., 15(1):99–102. http://dx.doi.org/10.1016/0378-1119(81)90108-6
Diatchenko, L., Lau, Y.F., Campbell, A.P., et al., 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. PNA., 93(12):6025–6030. http://dx.doi.org/10.1073/pnas.93.12.6025
Donnenberg, M.S., Kaper, J.B., 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun., 59(12):4310–4317.
Evans, P.M., Liu, C., 2005. SiteFind: a software tool for introducing a restriction site as a marker for successful site-directed mutagenesis. BMC Mol. Biol., 6(1):22. http://dx.doi.org/10.1186/1471-2199-6-22
Fushan, A.A., Drayna, D.T., 2009. MALS: an efficient strategy for multiple site-directed mutagenesis employing a combination of DNA amplification, ligation and suppression PCR. BMC Biotechnol., 9(1):83. http://dx.doi.org/10.1186/1472-6750-9-83
Gay, P., Le Coq, D., Steinmetz, M., et al., 1983. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J. Bacteriol., 153(3):1424–1431.
Georgi, T., Rittmann, D., Wendisch, V.F., 2005. Lysine and glutamate production by Corynebacterium glutamicum on glucose, fructose and sucrose: roles of malic enzyme and fructose-1,6-bisphosphatase. Metab. Eng., 7(4):291–301. http://dx.doi.org/10.1016/j.ymben.2005.05.001
Ho, S.N., Hunt, H.D., Horton, R.M., et al., 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gen., 77(1):51–59. http://dx.doi.org/10.1016/0378-1119(89)90358-2
Hogrefe, H.H., Cline, J., Youngblood, G.L., et al., 2002. Creating randomized amino acid libraries with the QuikChange® Multi Site-Directed Mutagenesis Kit. Biotechnique., 33(5):1158–1165.
Holland, E.G., Acca, F.E., Belanger, K.M., et al., 2015. In vivo elimination of parental clones in general and site-directed mutagenesis. J. Immun. Method., 417:67–75. http://dx.doi.org/10.1016/j.jim.2014.12.008
Homilton, C.M., Aldea, M.M., Washburm, B.K., et al., 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol., 171(9):4617–4622.
Horton, R.M., Cai, Z., Ho, S.N., et al., 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechnique., 8(5):528–535.
Hou, X.H., Chen, X.D., Zhang, Y., et al., 2012. L-Valine production with minimization of by-products’ synthesis in Corynebacterium glutamicum and Brevibacterium flavum. Amino Acid., 43(6):2301–2311. http://dx.doi.org/10.1007/s00726-012-1308-9
Hu, J., Li, Y., Zhang, H., et al., 2014. Construction of a novel expression system for use in Corynebacterium glutamicum. Plasmi., 75:18–26. http://dx.doi.org/10.1016/j.plasmid.2014.07.005
Ikeda, M., Katsumata, R., 1998. A novel system with positive selection for the chromosomal integration of replicative plasmid DNA in Corynebacterium glutamicum. Microbiology, 144(Pt 7):1863–1868. http://dx.doi.org/10.1099/00221287-144-7-1863
Imai, Y., Matsushima, Y., Sugimura, T., et al., 1991. A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res., 19(10):2785. http://dx.doi.org/10.1093/nar/19.10.2785
Imaizumi, A., Takikawa, R., Chie, K., et al., 2005. Improved production of L-lysine by disruption of stationary phase-specific rmf gene in Escherichia coli. J. Biotechnol., 117(1):111–118. http://dx.doi.org/10.1016/j.jbiotec.2004.12.014
Inui, M., Suda, M., Okina, S., 2007. Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiolog., 153(8):2491–2504. http://dx.doi.org/10.1099/mic.0.2006/005587-0
Johnston, C., Polard, P., Clavers, J.P., 2013. The DpnI/DpnII pneumococcal system, defense against foreign attack without compromising genetic exchange. Mobile Genet. Elem., 3(4):e25582. http://dx.doi.org/10.4161/mge.25582
Jones, D.H., Winistorfer, S.C., 1991. Site-specific mutagenesis and DNA recombination by using PCR to generate recombinant circles in vitro or by recombination of linear PCR products in vivo. Method., 2(1):2–10. http://dx.doi.org/10.1016/S1046-2023(05)80120-3
Judson, N., Mekalanos, J.J., 2000. Transposon-based approaches to identify essential bacterial genes. Trends Microbiol., 8(11):521–526. http://dx.doi.org/10.1016/S0966-842X(00)01865-5
Kalinowski, J., Bathe, B., Daniela, B., et al., 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J. Biotechnol., 104(1–3):5–25. http://dx.doi.org/10.1016/S0168-1656(03)00154-8
Karnik, A., Karnik, R., Grefen, C., 2013. SDM-Assist software to design site-directed mutagenesis primers introducing “silent” restriction sites. BMC Bioinformatic., 14(1): 105–114. http://dx.doi.org/10.1186/1471-2105-14-105
Kato, C., Ohimiya, R., Mizuno, T., 1998. A rapid method for disrupting gene in Escherichia coli genome. Biosci. Biotechnol. Biochem., 62(9):1826–1829. http://dx.doi.org/10.1271/bbb.62.1826
Ke, S.H., Madison, E.L., 1997. Rapid and efficient mutagenesis by single-tube “megaprimer” PCR method. Nucleic Acids Res., 25(16):3371–3372. http://dx.doi.org/10.1093/nar/25.16.3371
Khare, V., Eckert, K.A., 2002. The proofreading 3′-5′ exonuclease activity of DNA polymerases: a kinetic barrier to translesion DNA synthesis. Mutat. Res.-Fund. Mol. M., 510(1–2):45–54. http://dx.doi.org/10.1016/S0027-5107(02)00251-8
Kim, N.S., 2015. Transposable elements and genomics. Genes Genom., 37(2):111–112. http://dx.doi.org/10.1007/s13258-014-0262-1
Kim, Y.G., Maas, S., 2000. Multiple site mutagenesis with high targeting efficiency in one cloning step. Biotechnique., 28:196–198.
Kirsch, R.D., Joly, E., 1998. An improved PCR-mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res., 26(7):1848–1850. http://dx.doi.org/10.1093/nar/26.7.1848
Lacks, S., Greenberg, B., 1977. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J. Mol. Biol., 114(1):153–168. http://dx.doi.org/10.1016/0022-2836(77)90289-3
Li, J., Li, C.H., Xiao, W., et al., 2008. Site-directed mutagenesis by combination of homologous recombination and DpnI digestion of the plasmid template in Escherichia coli. Anal. Biochem., 373(2):389–391. http://dx.doi.org/10.1016/j.ab.2007.10.034
Liang, X.Q., Pen, L.S., Li, K., et al., 2012. A method for multisite-directed mutagenesis based on homologous recombination. Anal. Biochem., 427(1):99–101. http://dx.doi.org/10.1016/j.ab.2012.05.002
Liew, K.S., Ho, W.S., Pang, S.L., et al., 2015. Development and characterization of microsatellite markers in sawih tree (Duabanga moluccana Blume) using ISSR-suppression PCR techniques. Physiol. Mol. Biol. Plant., 21(1):163–165. http://dx.doi.org/10.1007/s12298-014-0262-2
Ling, M.M., Robinson, B.H., 1997. Approaches to DNA mutagenesis: an overview. Anal. Biochem., 254(2):157–178. http://dx.doi.org/10.1006/abio.1997.2428
Liu, H., Naismith, J.H., 2008. An efficient one-step sitedirected deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol., 8(1):91. http://dx.doi.org/10.1186/1472-6750-8-91
Lu, L., Patel, H., Bisser, J.J., 2002. Optimizing DpnI digestion conditions to detect replicated DNA. Biotechnique., 33(2):316–318.
Luo, F.G., Du, X.L., Weng, T.T., et al., 2012. Efficient multisite-directed mutagenesis directly from genomic template. J. Biosci., 37(Suppl. 1):965–969. http://dx.doi.org/10.1007/s12038-012-9257-8
Maloy, S.R., Nunn, W.D., 1981. Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol., 145(2): 1110–1112.
Mandaci, S., 2011. Site-directed mutagenesis as the cornerstone of protein engineering: from basic biotechnology to industrial enzymes. Curr. Opin. Biotechnol., 22(Suppl. 1): S39. http://dx.doi.org/10.1016/j.copbio.2011.05.093
Martens, J.H., Barg, H., Warren, M., et al., 2002. Microbial production of vitamin B12. Appl. Microbiol. Biotechnol., 58(3):278–285. http://dx.doi.org/10.1007/s00253-001-0902-7
Martin, A., Toselli, E., Rosier, M.F., et al., 1995. Rapid and high efficiency site-directed mutagenesis by improvement of the homologous recombination technique. Nucleic Acids Res., 23(9):1642–1643. http://dx.doi.org/10.1093/nar/23.9.1642
Meetei, A.R., Rao, M.R.S., 1998. Generation of multiple site-specific mutations in a single polymerase chain reaction product. Anal. Biochem., 264(2):288–291. http://dx.doi.org/10.1006/abio.1998.2866
Mitchell, L.A., Cai, Y.Z., Taylor, M., et al., 2013. Multichange isothermal mutagenesis: a new strategy for multiple site-directed mutations in plasmid DNA. ACS Synth. Biol., 2(8):473–477. http://dx.doi.org/10.1021/sb300131w
Montaldo, H.H., 2006. Genetic engineering applications in animal breeding. Electron. J. Biotechnol., 9(2):157–170. http://dx.doi.org/10.2225/vol9-issue2-fulltext-7
Muyrers, J.P.P., Zhang, Y.M., Stewart, A.F., 2001. Techniques: recombinogenic engineering—new options for cloning and manipulating DNA. Trends Biochem. Sci., 26(5): 325–331. http://dx.doi.org/10.1016/S0968-0004(00)01757-6
Nakashima, N., Miyazaki, K., 2014. Bacterial cellular engineering by genome editing and gene inactivation. Int. J. Mol. Sci., 15(2):2773–2793. http://dx.doi.org/10.3390/ijms15022773
Nossal, N.G., 1974. DNA synthesis on a double-stranded DNA template by the T4 bacteriophage DNA polymerase and the T4 gene 32 DNA unwinding protein. J. Biol. Chem., 249(17):5668–5676.
Patel, D.H., Wi, S.G., Bae, H.J., 2009. Modification of overlap expression PCR: a mutagenic approach. Indian J. Biotechnol., 8(2):181–186.
Peng, R.H., Xiong, A.S., Yao, Q.H., 2006. A direct and efficient PAGE-mediated overlap extension PCR method for gene multiple-site mutagenesis. Appl. Microbiol. Biotechnol., 73(1):234–240. http://dx.doi.org/10.1007/s00253-006-0583-3
Perlak, F.J., 1990. Single step large scale site-directed in vitro mutagenesis using multiple oligonucleotides. Nucleic Acids Res., 18(24):7457–7458. http://dx.doi.org/10.1093/nar/18.24.7457
Philippe, N., Alcaraz, J.P., Coursange, E., et al., 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmi., 51(3):246–255. http://dx.doi.org/10.1016/j.plasmid.2004.02.003
Picard, V., Ersdal-Badju, E., Lu, A., et al., 1994. A rapid and efficient one-tube PCR based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res., 22(13): 2587–2591. http://dx.doi.org/10.1093/nar/22.13.2587
Poustka, A., Rackwitez, H.R., Frischauf, A.M., et al., 1984. Selective isolation of cosmid clones by homologous recombination in Escherichia coli. PNA., 81(13):4129–4133. http://dx.doi.org/10.1073/pnas.81.13.4129
Qi, D., Scholthof, K.B.G., 2008. A one-step PCR-based method for rapid and efficient site-directed fragment deletion, insertion and substitution mutagenesis. J. Virol. Method., 149(1):85–90. http://dx.doi.org/10.1016/j.jviromet.2008.01.002
Reyrat, J.M., Pelicic, V., Gicquel, B., et al., 1998. Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infect. Immun., 66(9):4011–4017.
Saeedi, P., Moosaabadi, J.M., Sebtahmadi, S.S., et al., 2012. Site-directed mutagenesis in bacteriorhodopsin mutants and their characterization for bioelectrical and biotechnological equipment. Biotechnol. Lett., 34(3):455–462. http://dx.doi.org/10.1007/s10529-011-0731-4
Salerno, J.C., Jones, R.J., Erdogan, E., et al., 2005. A singlestage polymerase-based protocol for the introduction of deletions and insertions without subcloning. Mol. Biotechnol., 29(3):225–232. http://dx.doi.org/10.1385/MB:29:3:225
Sawitzke, J.A., Thomason, L.C., Bubunenko, M., et al., 2013. Recombineering: using drug cassettes to knock out genes in vivo. Methods Enzymol., 533:79–102. http://dx.doi.org/10.1016/B978-0-12-420067-8.00007-6
Schäfer, A., Tauch, A., Jäger, W., et al., 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutumicum. Gen., 145(1):69–73. http://dx.doi.org/10.1016/0378-1119(94)90324-7
Seraphin, B., Kandels-Lewis, S., 1996. An efficient PCR mutagenesis strategy without gel purification step that is amenable to automation. Nucleic Acids Res., 24(16): 3276–3277. http://dx.doi.org/10.1093/nar/24.16.3276
Seyfang, A., Jin, J.H., 2004. Multiple site-directed mutagenesis of more than 10 sites simultaneously and in a single round. Anal. Biochem., 324(2):285–291. http://dx.doi.org/10.1016/j.ab.2003.10.012
Shankarappa, B., Vijayananda, K., 1992. SILMUT: a computer program for the identification of regions suitable for silent mutagenesis to introduce restriction enzyme recognition sequences. Biotechnique., 12(6):882–884.
Siloto, R.M.P., Weselake, R.J., 2012. Site saturation mutagenesis: methods and applications in protein engineering. Biocatal. Agric. Biotechnol., 1(3):181–189. http://dx.doi.org/10.1016/j.bcab.2012.03.010
Siwek, W., Czapinska, H., Bochtler, M., et al., 2012. Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restriction endonuclease R. DpnI. Nucleic Acids Res., 40(15):7563–7572. http://dx.doi.org/10.1093/nar/gks428
Stoynova, L., Solórzano, R., Collins, E.D., 2004. Generation of large deletion mutants from plasmid DNA. Biotechnique., 36:402–406.
Sun, S.H., Huang, H., Qi, Y.C., et al., 2015. Complementary annealing mediated by exonuclease: a method for seamless cloning and conditioning site-directed mutagenesis. Biotechnol. Biotec. Eq., 29(1):105–110. http://dx.doi.org/10.1080/13102818.2014.988094
Suzuki, N., Okai, N., Nonaka, H., et al., 2006. Highthroughput transposon mutagenesis of Corynebacterium glutamicum and construction of a single-gene disruptant mutant library. Appl. Environ. Microbiol., 72(5):3750–3755. http://dx.doi.org/10.1128/AEM.72.5.3750-3755.2006
Tauch, A., Götker, S., Pühler, A., et al., 2002. The alanine racemase gene alr is an alternative to antibiotic resistance genes in cloning systems for industrial Corynebacterium glutamicum strains. J. Biotechnol., 99(1):79–91. http://dx.doi.org/10.1016/S0168-1656(02)00159-1
Tian, J., Liu, Q., Dong, S., et al., 2010. A new method for multisite-directed mutagenesis. Anal. Biochem., 406(1):83–85. http://dx.doi.org/10.1016/j.ab.2010.06.018
Tilly, K., Elias, A.F., Bono, J.L., et al., 2000. DNA exchange and insertional inactivation in spirochetes. J. Mol. Microbiol. Biotechnol., 2(4):433–442.
Tseng, W.C., Lin, J.W., Wei, T.Y., et al., 2008. A novel megaprimed and ligase-free, PCR-based, site-directed mutagenesis method. Anal. Biochem., 375(2):376–378. http://dx.doi.org/10.1016/j.ab.2007.12.013
Tu, H.M., Sun, S.S.M., 1996. Generation of a combination of mutations by use of multiple mutagenic oligonucleotides. Biotechnique., 20(3):352–354.
Turchin, A., Lawler, J.F., 1999. The primer generator: a program that facilitates the selection of oligonucleotides for site-directed mutagenesis. Biotechnique., 26(4):672–676.
Urban, A., Nenkirchen, S., Jaeger, K.E., 1997. A rapid and efficient method for site-directed mutagenesis using one-step overlap extension PCR. Nucleic Acids Res., 25(11):2227–2228. http://dx.doi.org/10.1093/nar/25.11.2227
Wan, H.S., Li, Y.W., Fan, Y., et al., 2012. A site-directed mutagenesis method particularly useful for creating otherwise difficult-to-make mutants and alanine scanning. Anal. Biochem., 420(2):163–170. http://dx.doi.org/10.1016/j.ab.2011.09.019
Wäneskog, M., Bjerling, P., 2014. Multi-fragment sitedirected mutagenic overlap extension polymerase chain reaction as a competitive alternative to the enzymatic assembly method. Anal. Biochem., 444:32–37. http://dx.doi.org/10.1016/j.ab.2013.09.021
Wang, H.P., Zhou, N., Ding, F., et al., 2011. An efficient approach for site-directed mutagenesis using central overlapping primers. Anal. Biochem., 418(2):304–306. http://dx.doi.org/10.1016/j.ab.2011.07.008
Wang, X.H., Pineau, C., Guschinskaya, N., et al., 2012. Cysteine scanning mutagenesis and disulfide mapping analysis of arrangement of GspC and GspD protomers within the Type 2 secretion system. J. Biol. Chem., 287(23): 19082–19093. http://dx.doi.org/10.1074/jbc.M112.346338
Weiner, M.P., Costa, G.L., Schoettlin, W., et al., 1994. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene, 151(1–2):119–123. http://dx.doi.org/10.1016/0378-1119(94)90641-6
Wu, D.G., Guo, X.W., Lu, J., et al., 2013. A rapid and efficient one-step site-directed deletion, insertion, and substitution mutagenesis protocol. Anal. Biochem., 434(2):254–258. http://dx.doi.org/10.1016/j.ab.2012.11.028
Xu, D.Q., Tan, Y.Z., Huan, X.J., et al., 2010. Construction of a novel shuttle vector for use in Brevibacterium flavum, an industrial amino acid producer. J. Microbiol. Method., 80(1):86–92. http://dx.doi.org/10.1016/j.mimet.2009.11.003
Xu, J.Z., Zhang, J.L., Guo, Y.F., et al., 2013. Improvement of cell growth and production of L-lysine by genetically modified Corynebacterium. glutamicum during growth on molasses. J. Ind. Microbiol. Biotechnol., 40(12): 1423–1432. http://dx.doi.org/10.1007/s10295-013-1329-8
Xu, J.Z., Han, M., Zhang, J.Z., et al., 2014a. Improvement of L-lysine production combines with minimization of by-products synthesis in Corynebacterium glutamicum. J. Chem. Technol. Biotechnol., 89(12):1924–1933. http://dx.doi.org/10.1002/jctb.4278
Xu, J.Z., Han, M., Zhang, J.Z., et al., 2014b. Metabolic engineering Corynebacterium glutamicum for the L-lysine production by increasing the flux into L-lysine biosynthetic pathway. Amino Acid., 46(9):2165–2175. http://dx.doi.org/10.1007/s00726-014-1768-1
Xu, J.Z., Xia, X.H., Zhang, J.Z., et al., 2014c. A method for gene amplification and simultaneous deletion in Corynebacterium glutamicum genome without any genetic markers. Plasmi., 72:9–17. http://dx.doi.org/10.1016/j.plasmid.2014.02.001
Xu, J.Z., Zhang, J.L., Guo, Y.F., et al., 2015. Genetically modifying aspartate aminotransferase and aspartate ammonia-lyase affects metabolite accumulation in L-lysine producing strain derived from Corynebacterium glutamicum ATCC13032. J. Mol. Catal. B Enzym., 113:82–89. http://dx.doi.org/10.1016/j.molcatb.2014.12.015
Xu, M.J., Zhang, R.Z., Liu, X.Y., et al., 2013. Improving the acidic stability of a β-mannanase from Bacillus subtilis by site-directed mutagenesis. Process Biochem., 48(8): 1166–1173. http://dx.doi.org/10.1016/j.procbio.2013.06.014
Yu, D.G., Ellis, H.M., 2000. An efficient recombination system for chromosome engineering in Escherichia coli. PNA., 97(11):5978–5983. http://dx.doi.org/10.1073/pnas.100127597
Zhang, X.L., Jantama, K., Moore, J.C., et al., 2007. Production of L-alanine by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol., 77(2):355–366. http://dx.doi.org/10.1007/s00253-007-1170-y
Zhang, Y.M., Buchholz, F., Muyrers, J.P.P., et al., 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet., 20(20):123–128. http://dx.doi.org/10.1038/2417
Zheng, L., Baumann, U., Reymond, J.L., 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res., 32(14):e115. http://dx.doi.org/10.1093/nar/gnh110
Zhou, S., Causey, T.B., Hasona, A., et al., 2003. Production of optically pure D-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl. Environ. Microbiol., 69(1):399–407. http://dx.doi.org/10.1128/AEM.69.1.399-407.2003
Author information
Authors and Affiliations
Corresponding author
Additional information
Project supported by the Natural Science Foundation of Jiangsu Province (No. BK20150149), the Fundamental Research Funds for the Central Universities (No. JUSRP51504), and the Youth Foundation of Jiangnan University (No. JUSRP115A19), China
ORCID: Jian-zhong XU, http://orcid.org/0000-0001-6555-6059
Rights and permissions
About this article
Cite this article
Xu, Jz., Zhang, Wg. Strategies used for genetically modifying bacterial genome: site-directed mutagenesis, gene inactivation, and gene over-expression. J. Zhejiang Univ. Sci. B 17, 83–99 (2016). https://doi.org/10.1631/jzus.B1500187
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.B1500187
Keywords
- Escherichia coli
- Corynebacterium glutamicum
- DNA manipulation
- Site-directed mutagenesis
- Gene inactivation
- Gene over-expression