Abstract
Fragile X-associated tremor/ataxia syndrome (FXTAS) is a neurodegenerative disorder associated with a premutation repeat expansion (55–200 CGG repeats) in the 5′ noncoding region of the FMR1 gene. Solitary intranuclear inclusions within FXTAS neurons and astrocytes constitute a hallmark of the disorder, yet our understanding of how and why these bodies form is limited. Here, we have discovered that FXTAS inclusions emit a distinct autofluorescence spectrum, which forms the basis of a novel, unbiased method for isolating FXTAS inclusions by preparative fluorescence-activated cell sorting (FACS). Using a combination of autofluorescence-based FACS and liquid chromatography/tandem mass spectrometry (LC-MS/MS)-based proteomics, we have identified more than two hundred proteins that are enriched within the inclusions relative to FXTAS whole nuclei. Whereas no single protein species dominates inclusion composition, highly enriched levels of conjugated small ubiquitin-related modifier 2 (SUMO 2) protein and p62/sequestosome-1 (p62/SQSTM1) protein were found within the inclusions. Many additional proteins involved with RNA binding, protein turnover, and DNA damage repair were enriched within inclusions relative to total nuclear protein. The current analysis has also allowed the first direct detection, through peptide sequencing, of endogenous FMRpolyG peptide, the product of repeat-associated non-ATG (RAN) translation of the FMR1 mRNA. However, this peptide was found only at extremely low levels and not within whole FXTAS nuclear preparations, raising the question whether endogenous RAN products exist at quantities sufficient to contribute to FXTAS pathogenesis. The abundance of the inclusion-associated ubiquitin- and SUMO-based modifiers supports a model for inclusion formation as the result of increased protein loads and elevated oxidative stress leading to maladaptive autophagy. These results highlight the need to further investigate FXTAS pathogenesis in the context of endogenous systems.
Similar content being viewed by others
Introduction
Fragile X-associated tremor/ataxia syndrome (FXTAS) is a progressive X-linked neurodegenerative disorder that arises from premutation CGG-repeat expansions (55-200 repeats) in the 5′ noncoding portion of the FMR1 gene [42]. The disorder, which generally has a clinical onset after age fifty, has core features of progressive cerebellar gait ataxia and kinetic tremor, with associated features of executive dysfunction, cognitive decline, neuropathy, dysautonomia, and Parkinsonism [14, 42, 45, 46]. Neuropathologic features of FXTAS include prominent white matter disease, loss of brain volume, Purkinje cell dropout, and solitary ubiquitin-positive inclusions within the nuclei of neurons and astrocytes [42, 44]. Inclusion formation is favored in cortical and hippocampal neurons and astrocytes and can be found in 2–20% of these cells in many patients [38]. Although many neurodegenerative diseases form inclusions and other aggregates in brain tissue, almost all of them include cytoplasmic aggregate formation and form multiple inclusions per cell [3, 7, 11, 17, 59]. Although FXTAS inclusions are found almost exclusively as solitary, spherical particles – distinct from nucleoli – within each nucleus, twinning of inclusions has been reported [5, 38, 39]. The mechanism(s) governing inclusion formation, and the nature of their composition, remain largely unknown; a better understanding of the properties of inclusions is likely to be key in understanding FXTAS pathogenesis.
FXTAS is largely limited to the premutation range, where there is normal to increased transcription of the expanded CGG-repeat mRNA [42, 68, 134]. The absence of the neurodegenerative phenotype for alleles in the full mutation range (>200 CGG repeats), with rare exceptions among mosaics [55, 82, 114], is thought to be due to methylation-coupled transcriptional silencing of the FMR1 gene. The requirement for transcriptional activity supports an RNA gain-of-function toxicity model [43, 78], as described earlier for myotonic dystrophy (DM) [25, 30, 89, 136].
Several specific models have been proposed to explain how neurotoxicity arises from the expanded CGG-repeat mRNA [118]. Analogous to the model for DM [136], the FXTAS mRNA sequestration model posits that the expanded-repeat FMR1 mRNA binds excessive amounts of one or more RNA-binding proteins [60, 115, 126, 127, 130], thus rendering those proteins functionally depleted. A second model proposes that initiation of translation at a non-AUG codon upstream of the CGG repeat generates an out-of-frame, toxic FMRpolyG protein [35, 125, 137]. Several sub-mechanisms are related to the FMRpolyG mechanism, including its inhibition of the ubiquitin-proteasomal system [53, 104] and co-aggregation with the nuclear lamina-associated polypeptide 2 beta (LAP2β) [125] or the splicing regulator transformer-2 protein homolog alpha (TRA2A) [19]. Previous studies have also found evidence for mediators of a DNA damage response (DDR) in both mouse and human tissues [44, 54, 58, 117]. However, the data of Robin et al. [117] suggest that the DDR may be a late response to early-onset calcium dysregulation and oxidative stress in affected neurons, progressing through the course of FXTAS pathogenesis. In support of this hypothesis, the FMR1 premutation model exhibits functional abnormalities early in development [16, 18, 23, 64, 81]. In all of these models, the role of inclusion formation/composition is of central importance, particularly with respect to its role as a repository for the products of aggregation/co-aggregation and to its use as a target for antibody staining to identify the proteins involved with FXTAS pathogenesis. Accordingly, determining both the general composition of the FXTAS inclusions and the estimated relative abundances of the component proteins is of critical importance.
Due to the technical challenges associated with purifying the FXTAS inclusions, their composition has been difficult to ascertain. Furthermore, the reliance on immunofluorescence methods to probe inclusion composition, and to direct their isolation, as in Iwahashi et al. [58], can introduce substantial bias because of the potential for antibody cross-reactivity and variability of the accessibility of their protein targets. In our efforts to address this issue, we have now observed that the FXTAS inclusions emit considerable autofluorescence across a broad range of wavelengths. By coupling FXTAS inclusion autofluorescence with fluorescence-activated cell sorting (FACS), we have been able to exploit this intrinsic property for rapid isolation of the inclusions for downstream characterization. We have coupled this unbiased autofluorescence-based isolation method with mass spectrometry (MS)-based proteomics to characterize the protein composition of FXTAS inclusions. Although using autofluorescence for sorting intracellular aggregates is new in the field of neurodegenerative disease, similar strategies have already been successfully applied in other fields [119, 129, 147]. This analysis allows us to estimate the relative abundances of certain proteins within FXTAS inclusions without the potential confounders of autofluorescence or antibody cross-reactivity, thus aiding the evaluation of existing models for FXTAS pathogenesis.
In the current work, we show that FXTAS inclusions are composed principally of protein and that, of the nearly 200 proteins that are enriched in the inclusions, over half are involved in RNA binding and/or protein turnover. In particular, heterogeneous nuclear ribonucleoproteins (hnRNPs), molecular chaperones, and protein modifiers are prevalent. No single protein species dominates the collection of enriched proteins. FMRpolyG is not detected in the FXTAS nuclei and is only detected at <0.05% molar abundance in the inclusions themselves. Among the most highly abundant of the enriched proteins are the small ubiquitin-related modifier 2 (SUMO 2), ubiquitin, and p62/sequestosome-1 (p62/SQSTM1; hereafter designated p62). Western blot and immunofluorescence experiments confirm that conjugated SUMO 2 is present at over 10 times higher levels in FXTAS patient brain nuclei compared with control brain nuclei and exists primarily in nuclear aggregates. SUMO 2 immunoprecipitation (IP) proteomics demonstrate that conjugated SUMO 2 in FXTAS samples is not due to conjugation to any one specific protein, but an overall higher level of conjugation to numerous proteins, specifically to DDR mediators and proteins involved in cellular response to oxidative stress. These results indicate that inclusions are mainly accumulations of proteins destined for removal that may have aggregated in response to the presence of RNA and/or exceeded the capacity of the nuclear proteasomal machinery. We suggest that once the size/abundance of the aggregates exceeds the threshold of proteasomal degradation, the continued aggregation leads to inclusion formation. For non-dividing cells, p62-directed autophagy is not available to clear the large nuclear aggregates, culminating in the trapped inclusion mass that is a hallmark feature of FXTAS.
Materials and Methods
Patient sample information and tissue preparation
Human postmortem frontal cortex samples from six FXTAS, one fragile X syndrome (FXS), one amyotrophic lateral sclerosis (ALS), one Parkinson’s disease (PD), and three control brains were obtained from the FXTAS brain repository at the University of California, Davis (UCD), School of Medicine. Additional human postmortem frontal cortex samples from two Alzheimer’s disease (AD), one frontotemporal dementia (FTD), two Huntington’s Disease (HD), and two progressive supranuclear palsy (PSP) cases were obtained from the NIH Brain & Tissue Repository. Human postmortem frontal cortex samples were also obtained from two AD cases through the UCD Alzheimer’s Disease Center. Tissue specimens were obtained through consented autopsies with their respective institutional review board approvals (UCD and VA West Los Angeles Medical Center). FXTAS patients had all been established through clinical diagnosis based on the presence of intention tremor, cerebellar ataxia, and parkinsonism, and were confirmed to have FXTAS based on the postmortem identification of intranuclear ubiquitin-positive inclusions in brain cells. Control tissue was obtained postmortem from individuals who did not have any significant neurological history, including encephalitis, epilepsy, demyelinating disease, dementia, or concurrent neurodegenerative disease. AD was rated using Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) criteria and Braak stage. All other brain tissues were neuropathologically examined and diagnosed according to clinical symptoms, gross features, and microscopic features. Samples were collected at the time of death and stored at -80 °C until used. Frozen tissue was dissected from the frontal cortex. High-premutation (hpCGG) and wild type (WT) mice with the C57BL/6 background were housed under standard vivarium conditions. All animal use protocols were approved by the Institutional Animal Care and Use Committee at UCD. Cerebral cortex was dissected and flash frozen in liquid nitrogen immediately after mice were sacrificed, then stored at -80 °C until used. Fibroblasts and lymphocytes used were collected and cultured as previously described in Pretto et al. [113]. All sample information is provided in Additional file 1: Tables S1-S3.
Immunofluorescence
Slides of brain nuclei were prepared using methods described previously [57, 58]. Slides were permeabilized in PBS + 0.5% Triton X-100 for 15 min at room temperature, then blocked in blocking buffer [3% bovine serum albumin (BSA) 0.5% Tween-20, in 1x PBS] for 30 min at room temperature. Primary antibody was diluted to desired concentration in detection buffer (1% BSA, 0.5% Tween-20, in 1x PBS), and slides were incubated in primary antibody for either 1 hr at room temperature or overnight at 4 °C in a humid chamber. After washing in PBS + 0.1% Tween-20, the slides were incubated in secondary antibody for 1 hr at room temperature in a humid chamber and stained with DAPI before coverslips were mounted with either SlowFade Antifade Mountant (for immediate viewing) or Prolong Diamond Antifade Mountant (for longer term viewing or storage). For each immunofluorescence experiment, secondary antibody-only slides and DAPI-only slides were prepared for comparison. Immunofluorescence primary antibodies: rabbit anti-SUMO 2/3 (Abcam ab3742; diluted 1:200), mouse anti-SUMO 2/3 (Abcam ab81371; diluted 1:500), rabbit anti-p62/SQSTM1 (Invitrogen 701510; diluted 1:100), mouse anti-ubiquitin (Bio-rad MCA-1398G; diluted 1:200), rabbit anti-ubiquitin (Abcam 134953; diluted 1:100). Secondary antibodies: goat anti-rabbit Alexa-647 (Abcam ab150083; diluted 1:500), goat anti-mouse Alexa-647 (Abcam ab150115; diluted 1:500), goat anti-mouse Alexa-488 (Abcam ab150113; diluted 1:500), goat anti-rabbit Alexa-488 (Abcam ab150077; diluted 1:500), goat anti-rabbit Alexa-532 (Invitrogen A-11009; diluted 1:500).
DNA fluorescence in situ hybridization (FISH)
To generate DNA FISH probes, biotin-labeled PCR amplicons corresponding to unique regions spanning and surrounding the FMR1 gene were generated. Metaphase slides were prepared from primary patient fibroblasts by applying colcemid to fibroblasts at ~70% confluency for 3 hr before cells were collected by trypsinization. Cells were incubated in KCl buffer before fixation using 3:1 methanol acetic acid. 0.3-0.5 μg of probe per slide was applied overnight to cells that were permeabilized successively in several buffers containing detergents, formamide, and ethanol. Bound biotin probe was incubated with Streptavidin bound to Alexa-555 (Invitrogen S32355) at a 1:200 dilution before staining with DAPI and mounting with Prolong Diamond Antifade Mountant.
Inclusion fractionation
Continuous sucrose gradients were prepared and frozen beforehand using sucrose solutions at concentrations of 2.6, 2.5, 2.4, 2.3, and 2.2 M. In thin-walled polyallomer centrifuge tubes, 6 ml of each solution was added and subsequently submerged in a dry ice ethanol bath, starting with the most concentrated at the bottom and progressing upwards to the least concentrated at the top. Frozen gradients were stored at -20 °C.
Nuclear isolation was performed on frozen human cortical tissue using modified methods from Iwahashi et al. [57] and McEwen and Zigmond [94]. Nuclei were centrifuged and resuspended in 800 μl of BDC + NP-40 [40 mM Tris, 10 mM NaCl, 10 mM CaCl2, 5 mM MgCl2, 0.1% NP-40, Complete protease inhibitor (Roche), RNase inhibitor (NEB), pH 7.9] and 200 U/ml of DNase I (NEB). Nuclei were Dounce-homogenized with a tight pestle on ice for 30 strokes, after which, the sample was rotated at 37 °C for 6 hr with periodic mixing by pipetting to reduce viscoelasticity. An aliquot was removed at this point to serve as total nuclear protein samples for MS. The remaining sample was centrifuged at 16,000 RCF for 10 min at 4 °C, and the pellet containing FXTAS inclusions was resuspended in BCC (20 mM HEPES, 400 mM NaCl, 1 mM DTT, 1 mM EDTA, 1 mM EGTA, Complete protease inhibitor, pH 7.4) with RNase inhibitor and frozen at -80 °C overnight.
Pre-made sucrose gradients were taken from -20 °C and placed at 4 °C overnight to allow thawing. The frozen, resuspended inclusion samples were thawed on ice, centrifuged at 16,000 RCF for 10 min at 4 °C, then diluted to 10 ml of BCC and carefully pipetted onto the top surface of the sucrose gradient, followed by ultracentrifugation at 100,000 RCF for 6 hr at 4 °C. 2 ml fractions were collected through a small needle puncture at the bottom of each centrifuge tube. Each fraction was diluted with BCC, centrifuged at 3,000 RCF for 10 min at 4 °C, then resuspended in BCC + RNase inhibitor and stored at -80 °C until FACS processing. Slides were made of each fraction to confirm presence of inclusions at fractions corresponding to densities of approximately 1.30 g/ml.
Western blot
Concentrations of protein lysates were measured using the Pierce BCA or MicroBCA assay kit (Thermo Scientific), and either 10 μg or 20 μg of protein per sample was mixed with 1x Tris buffered saline (TBS) and Laemmli buffer (375 mM Tris-HCl, 6% SDS, 4.8% glycerol, 9% 2-Mercaptoethanol, 0.01% bromophenol blue) to obtain a total volume of 30 μl. Samples were held at 95 °C for 5–10 min and allowed to cool to room temperature before loading onto Criterion TGX 18-well Any kD polyacrylamide gels (Bio-rad) alongside the Chameleon Duo protein ladder (Li-Cor). Samples were run at 20 mA for 20 min until clearly stacked in the gel, then run at 80 mA for 45 min. Gel was then transferred overnight at 4 °C onto a nitrocellulose membrane. The membranes were stained for total protein using Revert Total Protein Stain (Li-Cor) and imaged immediately for total protein on a Li-Cor Odyssey Imager before incubating in blocking buffer (5% BSA in 1x TBS) at room temperature for 1 hr. Primary antibodies were diluted in detection buffer (5% BSA + 0.1% Tween-20 in 1x TBS) and membranes were incubated in primary antibody overnight at 4 °C. The membrane was washed three times in wash buffer (0.1% Tween-20 in 1x TBS) before incubating in secondary antibody diluted in detection buffer at room temperature for 1 hr. Membranes were again washed three times in wash buffer and once in 1x TBS before imaging on a Li-Cor Odyssey Imager. Image processing and densitometry was performed using ImageStudio software. Specific protein signal was normalized both to adjacent background on the image and to total protein signal for each lane. Western blot primary antibodies: rabbit anti-SUMO 2/3 (Abcam ab3742; diluted 1:1,000), rabbit anti-p62/SQSTM1 (Invitrogen 701510; diluted 1:500). Secondary antibody: IRDye 800CW donkey anti-rabbit (Li-Cor; diluted 1:20,000).
Fluorescence-activated cell sorting (FACS) purification of inclusions
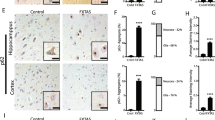
To sort inclusions for MS, two FXTAS patients (cases B3 and B6 from Table S1) were chosen for analysis alongside one control sample (case B8 from Table S1). Frozen inclusion-enriched fractions of sucrose density gradients from FXTAS nuclei, and equivalent density fractions from unaffected individuals, were thawed and immediately assessed for particle scatter and intrinsic autofluorescence characteristics by flow cytometry using a Beckman Coulter MoFlo Astrios EQ cell-sorting flow cytometer. As observed by fluorescence microscopy (Fig. 1 a), the inclusions present in FXTAS tissue homogenates were small, relatively homogenous in size, and primarily exhibited green autofluorescence (500–565 nm) following 488 nm laser excitation. These green fluorescent particles were not apparent in similarly prepared samples from control tissues. As is standard practice for detection of small subcellular particles [109], we used logarithmic scaling to distinguish inclusions from debris artifacts introduced in the sample buffer. We then removed larger aggregates by plotting the duration of 90° laser light scatter to remove objects with markedly increased laser dwell rates relative to the shorter transit times of single particles. Using these settings, we compared the strength of the autofluorescence signals in FXTAS and control samples across several detectors. We noted that the strongest fluorescence signal was measured in the green detector from 488 nm laser excitation, but this signal was markedly diminished for ≥670 nm wavelengths when subjected to 488, 561, or 640 nm laser excitation. In total, 8.6 million inclusions were sorted from one FXTAS patient sample and 6.5 million inclusions were sorted from a second FXTAS patient sample. Sorted inclusions were centrifuged at 3,000 RCF for 1 hr at 4 °C, and the pellets were pooled for each patient and resuspended in PBS. Aliquots of all samples were taken for the MicroBCA assay. SDS was added to samples to a final concentration of 5% to dissolve insoluble material. Sorted inclusion samples did not contain any visible precipitate after samples were mixed in SDS. MicroBCA assay kits were used according to the manufacturer’s instructions. Sorted inclusion samples yielded comparable total protein concentrations, at 50 μg/ml for the sample containing 6.5 million inclusions, and 68 μg/ml for the sample containing 8.6 million inclusions. Based on these concentrations, inclusions contain approximately 4x10-6 μg of protein per inclusion. BCA estimates were used to obtain 25 μg of protein from each sample for MS analysis.
a,c Flow-sorted FXTAS inclusions exhibit the same size and autofluorescent properties as in situ FXTAS inclusions. FXTAS inclusions exhibit strong autofluorescence at 480 nm and 545 nm, and weak autofluorescence at 360 nm and 620 nm wavelengths. Inclusions sorted by flow cytometry were verified by microscopy to confirm that sorted inclusions exhibit no significant difference from FXTAS inclusions viewed in situ. Slides were stained with DAPI only and viewed at 100x. Orange arrows denote inclusions, upper left labels indicate the wavelength used for the image, and upper right labels indicate the exposure level used to take the image. No postprocessing adjustments were made to brightness/contrast. b Fractions enriched in inclusions and submitted for FACS contain a population of FXTAS-specific particles identified by size and fluorescence properties. Logarithmic scaling was used on the detectors assigned to laser light scatter measurements (Inclusion scatter), and larger aggregates were removed by plotting the duration of 90° laser light scatter to remove objects with markedly increased laser dwell rates relative to the shorter transit times of single particles (Single inclusions). Sorted particles were identified as a population in FXTAS samples that was absent in control samples which exhibited strong green fluorescence emission and weak red fluorescence emission (gates in Green autofluorescence and Red autofluorescence, respectively). d Inclusions sorted sequentially by nuclear isolation, sucrose gradient, and flow cytometry are of high concentration and purity. Sorted inclusion samples viewed at 60x show high purity, with 80-90% of autofluorescent particles displaying autofluorescent properties consistent with FXTAS inclusions. 10-20% of particles (orange arrows) display a high level of autofluorescence in the far-red wavelengths, indicating that a small degree of non-inclusion debris may be present. Scale bars = 5 μm
SUMO 2/3 IP from nuclear lysate
A total of 6 g of frozen frontal cortical tissue, previously powdered under liquid nitrogen, was processed from two FXTAS patients and two age-matched control patients (FXTAS samples B2 and B4 and control samples B7 and B8 from Table S1). Nuclear isolation was performed as described above, except 1 mM EDTA, 1 mM EGTA, and 50 mM N-ethylmaleimide (Millipore Sigma) was added to the buffers. Isolated nuclei were lysed, passed through a 27-gauge needle, heated, then diluted before being centrifuged to remove any remaining insoluble material. Each sample was incubated with SUMO 2/3 antibody for 1 hr at 4 °C with rotation, and Protein G Magnetic Beads (NEB) were incubated with lysate-antibody mixture for 3 hr at 4 °C with rotation. Beads were then placed on a magnetic rack and the supernatant was collected to be run on western blot as the nonbound fraction. Approximately 1/10 of the beads from each IP was separated and eluted using urea elution buffer (7 M urea, 20 mM Tris, pH 7.5, 100 mM NaCl) to be run on western blots as IP elute. The remaining beads were washed three times with 50 mM triethylammonium bicarbonate (TEAB) buffer before on bead digestion.
Proteomics sample preparation
For proteomic analysis of isolated inclusions and total nuclear protein, sample eluates were dried down in a Centrivap centrifugal vacuum concentrator (Labconco, Kansas City, USA) and reconstituted in 50 μl of SDS solubilization buffer (5% SDS, 50 mM TEAB, 1X PhosSTOP phosphatase and Complete mini protease inhibitor tabs). Samples were clarified by centrifugation at 20,000 RCF for 10 min, supernatants set aside, and the resulting pellet further solubilized with 45 μl 100% formic acid for 1 hr at 37 °C. Solubilized pellets were dried down and recombined with soluble supernatants for analysis. Each volume of normalized sample was enzymatically digested with trypsin using S-Trap micro (Protifi, Huntington, NY) spin columns according to manufacturer instructions with the following modifications: samples were reduced with 20 mM DTT (Millipore Sigma) for 30 min at 56 °C, alkylated with 40 mM IAA (Millipore Sigma) for 30 min at room temperature, and trypsin (Worthington, Lakewood, NJ, USA) was added at a 1:12.5 ratio (enzyme (μg): protein (μg)) and reacted for 2 hr at 47 °C.
For FMRpolyG-GFP heterologous expression samples, harvested cell pellets were lysed directly in 200 μl SDS solubilization buffer with sonication (Qsonica Q125, Newton, CT; 2 rounds of alternating 10 sec on/10 sec off at 20% amplitude). For each sample, protein concentration was determined by BCA assay, volume was normalized to 140 μg of total protein, reduced and alkylated, and enzymatically digested with trypsin using S-Trap mini (Protifi) spin columns according to manufacturer instructions with the following modifications: samples were reduced with 20 mM DTT for 10 min at 50 °C, alkylated with 40 mM IAA for 30 min at room temperature, and two rounds of trypsin were added at a 1:25 ratio (enzyme (μg): protein (μg)) and reacted first for 2 hr at 37 °C followed by a second round overnight at 37 °C.
Sumo-IP samples were on-bead digested with 36 μg trypsin overnight at 37 °C in 50 mM TEAB without reduction or alkylation. Supernatants containing digested peptides were acidified to 1% TFA final concentration. All resulting sample elutes were dried and reconstituted in 2% acetonitrile/0.1% TFA for LC-MS/MS analysis.
LC-MS/MS and Data Analysis
Digested peptides were analyzed by LC-MS/MS on a Thermo Scientific Q Exactive Plus Orbitrap Mass Spectrometer (Waltham, MA, USA) in conjunction with an EASY-nLC 1200 UHPLC and Proxeon nanospray source operating in positive ionization mode. Peptides were loaded on a 100 μm x 25 mm Magic C18 100Å 5U reverse phase trap before being separated using a 75 μm x 150 mm Magic C18 200Å 3U reverse phase column. Peptides were eluted with an increasing percentage of acetonitrile over the course of a 90- or 120-min gradient with a flow rate of 300 nl/min. An MS survey scan was obtained for the m/z range 350–1600 and acquired with a resolution of 70,000 and a target of 1x106 ions or a maximum injection time of 30 msec. MS/MS spectra were acquired using a top 15 method where the top 15 ions in the MS spectra were subjected to high energy collisional dissociation. MS/MS spectra were acquired with a resolution of 17,500 and a target of 5x104 ions or a maximum injection time of 50 msec. An isolation mass window of 1.6 m/z was used for precursor ion selection, charge states of 2-4 were accepted, and a normalized collision energy of 27% was used for fragmentation. A 20-sec duration was used for dynamic exclusion. For identification and quantitation of data-dependent acquisition data, raw files were searched with Andromeda in MaxQuant version 1.6.1.0 and further processed in Perseus version 1.6.0.2 or Scaffold version 4.8.4.
Additional details for all methods are provided in Additional file 1: Methods.
Results
Intrinsic autofluorescence of human FXTAS inclusions facilitates their isolation using preparative flow cytometry
During a series of immunofluorescence studies of FXTAS inclusion-bearing frontal cortex, we observed that the inclusions emit broad-spectrum autofluorescence. Inclusion autofluorescence has previously been reported in neuronal intranuclear inclusion disease (NIID) [70, 110], but has not been reported previously for FXTAS inclusions. Examination of slides generated from multiple FXTAS patients revealed that the inclusions emit the lowest intensity of autofluorescence at 360 nm and 620 nm, with maximal brightness by visual inspection between 480 nm and 545 nm (Fig. 1 a). Stimulated emission depletion (STED) microscopy on a Leica SP8 STED 3x was used to measure the excitation/emission spectra of FXTAS inclusions, and the results corroborated what was seen on visual examination (Additional file 1: Figure S1a), with maximum emission intensity at 550 nm by STED. Autofluorescence has been found in every FXTAS case examined thus far (~10 cases), with spectral properties quantitatively consistent across samples. Additional experiments were performed to ensure that inclusion autofluorescence is not an artifact of fixation, mounting, or staining (unpublished data).
The constancy of FXTAS inclusion autofluorescence has enabled us to isolate relatively pure samples of inclusions using preparative flow cytometry. To minimize contamination due to the known autofluorescence of lipofuscin [27], we isolated nuclei from frozen postmortem cortical tissue by a modification of previous methods [58, 83, 94], followed by sucrose gradient fractionation of disrupted nuclei to obtain a crude inclusion fraction. Our approach, which substantially removes cytoplasmic lipofuscin during the nuclear isolation, largely eliminates non-inclusion autofluorescent cellular material. Ultracentrifugation of nuclear homogenate on a sucrose gradient produces a diffuse band of particles at a density of ~1.30 g/ml, visible in FXTAS samples but not in control samples (Additional file 1: Figure S1b). Visualization of the particles within this band by fluorescence microscopy reveals autofluorescent particles that exhibit the same spectral properties as in situ FXTAS inclusions. However, this inclusion-enriched isolate still contains a substantial fraction of non-inclusion particles.
To obtain inclusion samples of greater purity, preparative FACS was used to sort inclusions based on their intrinsic autofluorescence (i.e., without the use of antibodies). The population of particles in the 1.30 g/ml banded material was sorted using size and autofluorescence characteristics (Fig. 1 b). Sorted populations of particles viewed by microscopy exhibit the same spectral properties as in situ FXTAS inclusions (Fig. 1 c). FACS-sorted inclusions can be centrifuged and resuspended in a smaller volume to obtain a concentrated sample of relatively pure inclusions (Fig. 1 d). Although most of the particles in these sorted samples exhibit the same spectral properties as inclusions, some contaminants appear too bright at 620 nm to be considered as inclusions and are likely lipofuscin particles that were not completely removed during the nuclear isolation process. Taking these into account, we estimate that FACS sorting inclusion preparations are of 80–90% purity.
FXTAS inclusions appear to be comprised mainly of protein, with a smaller component of RNA, but do not contain significant amounts of DNA
Inclusion-enriched sucrose fractions (1.30 g/ml density) were used to provide a crude estimate of the general molecular composition of inclusions. The inclusions were treated with either DNase I, RNase A/T1, Proteinase K, or no enzyme; each sample was viewed by fluorescence microscopy utilizing autofluorescence (no immunostaining). The no-enzyme and DNase I treatments exhibited wholly intact inclusions, with no diminishment of autofluorescence intensity, inclusion size, or integrity, indicating that DNA does not comprise a significant proportion of the makeup of inclusions (Fig. 2). In comparison, RNase treated inclusions displayed substantial loss of autofluorescence intensity, with many of the inclusions, while still present, appearing to have lost structural integrity. This observation suggests that RNA may play a significant role in the structural makeup of the inclusions, which supports previous evidence that FMR1 mRNA is present in inclusions [135]. However, preliminary attempts to analyze the RNA complement through RNAseq, although confirming the presence of mRNA, were confounded by extensive degradation of the RNA isolated from the inclusions. Further efforts to determine the composition of the RNA species within the inclusions are ongoing. Finally, Proteinase K treatment alone completely abolished visualization of any inclusions, suggesting that the inclusions are predominantly proteinaceous.
FXTAS inclusions are largely made up of protein and contain a smaller component of RNA, but do not contain significant amounts of DNA. DNase treatment did not affect the quantity, integrity, or level of autofluorescence exhibited by isolated inclusions, detected at 488nm, 555nm; not visualized at 647nm without several fold increase in brightness. RNase treatment did not affect the quantity of inclusions seen, but the autofluorescence levels exhibited by inclusions was decreased. Postprocessing to increase the brightness of RNase treated inclusions revealed that the integrity of the inclusions appeared compromised. Protease treatment completely abolished visualization of inclusions using autofluorescence, and no inclusions were observed. Unless specified (bright), all images were acquired for 208.1 ms
FXTAS inclusions are primarily heterogeneous protein aggregates enriched for proteins involved in RNA binding and protein turnover and degradation
MS-based proteomics was utilized to determine the protein composition of FXTAS inclusions purified from frontal cortex of two FXTAS patients (cases B3 and B6 from Table S1) with high loads of inclusions (~9–14% inclusion-bearing neuronal/astrocytic cells), as previously determined through immunohistochemistry. Patient samples were processed by nuclear isolation/sucrose gradient centrifugation/preparative FACS to obtain sorted inclusion samples. These samples were compared by LC-MS/MS to total nuclear protein from the same samples, as well as to the nuclear isolate from a control non-FXTAS sample (case B8 from Table S1). In general, there was no dominant protein among the inclusion-enriched proteins (see full MS dataset in Additional file 1). Although histones comprised 25–70% of each inclusion protein isolate, none of the histone species was enriched in inclusions over total nuclear protein. The next highest abundance protein, ubiquitin, only comprises ~5% of the inclusion protein complement. To identify proteins likely to be specifically involved in inclusion formation, only those proteins whose molar fractions equaled or exceeded 0.01% of the total inclusion protein content for both sorted inclusion samples and showed at least 1.5-fold enrichment in both sorted samples over their accompanying total nuclear samples were considered for further analysis. One-hundred seventy-six proteins fit these criteria out of a total of ~1,900 proteins identified by MS analysis of the purified inclusions (Additional file 2). The enriched proteins were scored and categorized by function according to UniProt. Three major functional categories stood out among the enriched proteins: RNA binding proteins, proteins involved in protein turnover, and DNA binding proteins (Fig. 3).
Categorization by function of inclusion-enriched proteins reveals large populations of proteins involved in RNA binding and protein turnover. Proteins that were found present to at least 0.01% in both FXTAS sorted inclusion samples and showed at least 50% enrichment in sorted inclusion samples over accompanying total nuclear samples were identified for categorization. A total of 176 proteins fit these criteria. Proteins were scored by main function(s) as identified by UniProt, and each protein could have multiple scores. Percentages were calculated as number of proteins with that score out of the total number of scores given in the dataset. The “Other” category contains proteins involved in functions including biosynthetic processes, energy metabolism, immunity, and neural development, with no one single category exceeding 3%. A total of 75 proteins were scored as RNA binding proteins and proteins participating in RNA metabolism (transcription, editing, splicing, transport). Many of the higher abundance proteins in this category were hnRNPs. Although there were many ribosomal proteins found in this dataset, almost all of them were found at low abundance (<0.1% of the total protein composition) and were not found to have more total nuclear enrichment in FXTAS nuclei compared to control nuclei. We hypothesize that these proteins represent mainly background through the isolation process and are present more as bystanders rather than main players in inclusion formation. A total of 32 proteins were scored as those participating in protein turnover (protein folding, aggregation, modification, degradation). Within this category, the majority of protein species are those involved in binding and processing proteins destined for recycling or removal. The protein species comprising this category have the highest abundance levels in inclusions. A total of 21 proteins were scored as DNA binding proteins or proteins participating in DNA metabolism (chromatin remodeling, replication, repair). Of these, 6 are players in DNA damage repair, including RAD50, RPA1, and XRCC6
Over one-third of the inclusion-enriched proteins are RNA binding proteins. Within this category, 17.3% are ribonucleoproteins. Many of these ribonucleoprotein species (mainly hnRNPs) are present at relatively high abundance within the enriched fraction, half making up over 0.2% in both inclusion samples, and two, hnRNPA1 and hnRNPA3, comprising 1-2% of both inclusion samples. A further 26.7% of the inclusion-enriched RNA binding proteins are ribosomal proteins. However, almost all of these species were present at low abundance (<0.1% of total inclusion protein). They also were not found at higher levels in FXTAS total nuclear samples compared with control total nuclear samples. We also detected approximately forty proteins involved with RNA splicing, the most abundant being U2AF (~0.2%) and SFPQ (~0.5%); both are slightly enriched in the inclusion fraction (by 2.0- and 1.4-fold, respectively). However, there does not appear to be any overall trend for enrichment, with the majority of detected splicing factors being slightly less abundant in the inclusions than in the surrounding nuclear matrix.
A further 15% of inclusion-enriched proteins were scored as involved in protein turnover; that is, proteins that are tasked with binding and processing of other proteins for recycling or removal. Although the protein turnover category contains fewer members compared to the RNA binding category, the protein species comprising this category have the highest abundance levels in inclusions. Within this category, 40% of members were categorized as molecular chaperones, 22% play roles in protein modification, and 22% were members of proteasomal machinery.
The final major functional category, DNA binding proteins, makes up 10% of the population of inclusion-enriched proteins, and 28.6% of these were categorized as DDR mediators. Among these DDR mediators were proteins such as RAD50, RPA1, and XRCC6.
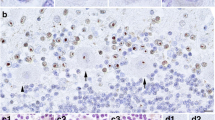
For a more focused analysis of the highest-abundance inclusion-enriched proteins, those proteins which make up at least 0.5% of a sorted inclusion sample and which were enriched by at least 50% were considered further. Only 15 proteins fit these criteria (Table 1), with the most prominent members being ubiquitin and SUMO 2. Other proteins in this list include several hnRNPs and chaperones. Only five proteins were enriched by at least three-fold in both sorted inclusion samples: ubiquitin, SUMO 2, myeloid leukemia factor 2 (MLF2), myelin basic protein (MBP), and p62. Western blot to confirm MS results for p62 was performed on five replicates of control versus FXTAS brain nuclear lysates from control (n=3) and FXTAS (n=4) patients. p62 was seen at 62 kDa and was approximately three-fold more intense in FXTAS samples compared with control samples (Additional file 1: Figure S2a), and immunofluorescence on FXTAS brain smears confirmed the presence of p62 in nuclear inclusions (Additional file 1: Figure S2b).
Fold change values for each of the FXTAS inclusion samples over their accompanying FXTAS nuclear samples and the control nuclear sample are shown, with the abundance values as a percentage of the total identified protein molar composition for each sample displayed in parentheses. Proteins identified were present in at least one FXTAS sorted inclusion sample to at least 0.5% and showed at least 50% enrichment in both FXTAS sorted inclusion samples over corresponding FXTAS nuclear samples. Only five of these proteins (bolded) were enriched at least 3-fold in FXTAS inclusions over accompanying FXTAS total nuclear proteins, and only SUMO 2/3, MLF2, and p62 were also found to be enriched at least 3-fold in FXTAS total nuclear samples over control total nuclear sample. Numeric values presented outside parentheses are fold change of inclusion samples over nuclear samples. Values inside parentheses show the abundance values as a percentage of the total protein composition identified for inclusion samples over nuclear samples.
SUMO 2/3 conjugates are present in FXTAS brain nuclei at levels exceeding ten-fold over the levels found in brain nuclei from controls
To confirm elevation of SUMO 2 in FXTAS brain nuclei, western blot analysis was performed using a SUMO 2/3 antibody on protein lysates from whole brain tissue and nuclear brain protein lysates from FXTAS (n=5) patients and controls (n=3) (Fig. 4 a, Additional file 1: Figure S3). Although MS analysis can distinguish SUMO 2 from SUMO 3, most antibodies recognize both species due to their high level of sequence similarity. Each FXTAS nuclear sample exhibited an intense SUMO 2/3 smear extending from the well down to around 25–30 kb, indicating an unusually large amount of conjugated SUMO 2/3, especially at higher molecular weights. This smear was not apparent in whole brain tissue lysates, indicating that the SUMO conjugates are present primarily or exclusively in the nuclear compartment, with the conjugates being diluted in whole tissue. Quantification revealed an over ten-fold elevation of conjugated SUMO 2/3 in FXTAS nuclei compared to control nuclei. Interestingly, no significant differences were seen between FXTAS and control for the SUMO 1 isoform (Additional file 1: Figure S4).
a FXTAS nuclear protein samples exhibit over ten times higher SUMO 2/3 protein levels compared to control nuclear protein samples. FXTAS nuclear protein samples exhibit a vibrant SUMO 2/3 smear indicating over 10 times higher levels of conjugated SUMO 2/3 over control nuclear protein samples after normalizing to total protein levels (unpaired student’s t-test (single group) with p-value <0.005). Whole tissue sample signals were extremely faint by comparison. No significant difference was seen between control and FXTAS samples for unconjugated SUMO 2/3. b SUMO 2/3 immunofluorescence reveals that SUMO 2/3 is present mainly as large, concentrated intranuclear aggregates in FXTAS brain cells, and FXTAS inclusions always colocalize with SUMO 2/3 aggregation. SUMO 2/3 forms large, intranuclear aggregates in FXTAS brain cells that colocalize with FXTAS inclusions. Immunofluorescence was performed using an Alexa 647 secondary antibody to minimize the effect of inclusion autofluorescence. 360nm+480nm images show inclusion autofluorescence. In FXTAS samples, SUMO 2/3 aggregates at 620nm are so vibrant that the signal intensity at 208.1ms is distorted, and the signal is bright even at an exposure of 10.1ms. No post-processing was done to alter brightness/contrast levels. Orange arrows denote FXTAS inclusions. When looking at clusters of nuclei (bottom panels), control brain cells show diffuse SUMO 2/3 staining throughout nuclei, but no aggregates, whereas FXTAS brain cells show large, circular aggregates in a large proportion of cells. c SUMO 2/3 aggregates are present in mouse high premutation CGG brain nuclei but are not present in wildtype brain nuclei. Intranuclear SUMO 2/3 aggregates are also seen in 11-month-old premutation CGG male mouse brain, but not in age-matched male wildtype mice. Aggregates were smaller and fewer than those seen in human FXTAS brain. Immunofluorescence was performed using an Alexa-647 secondary antibody. No postprocessing was done to alter brightness/contrast levels. Orange arrows denote SUMO 2/3 aggregates. d The SUMO 2/3 smears found in FXTAS nuclear protein samples are not present to the same degree in other neurodegenerative brain samples investigated. Western blot analysis was performed on nuclear lysates from human frontal cortex tissue obtained from a variety of neurodegenerative disorders. Due to the lack of availability of sufficient material, replicates were not possible for several samples. Where replicates were possible, error bars are shown. Scale bars = 10 μm
Immunofluorescence of control and FXTAS brain nuclei was used to localize the elevated SUMO 2/3. Staining was performed in the far-red spectral region (620 nm) where autofluorescence is minimal; secondary antibody-only controls are shown for comparison. SUMO 2/3 staining produced an intensely bright signal within inclusions (Fig. 4 b, top panels). All inclusions that were identified by autofluorescence at 480 nm stained intensely positive for SUMO 2/3 at 620 nm. In addition, bright SUMO 2/3 aggregates found at 620 nm could be used to identify smaller inclusion-like aggregates that were not immediately apparent at 480 nm or 545 nm. This suggests that SUMO 2/3 accumulation may be an early and major participant in inclusion formation. Outside of the SUMO 2/3 aggregates, there did not appear to be any generalized increase in SUMO signal in the rest of the nucleus compared to control. The quantity of SUMO-staining aggregates became apparent when looking at incompletely homogenized clumps of cells in control versus FXTAS brain, where SUMO staining only lightly and diffusely stained the cells in control, whereas many SUMO 2/3 aggregates were present in FXTAS (Fig. 4 b, bottom panels).
The same analysis of SUMO 2/3 immunofluorescence was performed on 11-month-old, high premutation CGG (hpCGG; ~170 CGG repeats), and wildtype (WT) mouse brain tissue to determine whether SUMO 2/3 aggregation also occurs in an FMR1 premutation mouse model [10, 142]. Many SUMO 2/3 aggregates were apparent in both male and female homozygous hpCGG mouse brain but were not present in age-matched WT mice (Fig. 4 c, Additional file 1: Figure S5a). However, the number and size of the aggregates were smaller than those present in human brain. Western blot analysis of hpCGG mouse brain nuclear lysates did not exhibit the same prominent SUMO 2/3 smears present in human FXTAS brain, perhaps because the premutation mice were still in the premutation phase (only 11 mo old) whereas FXTAS patient postmortem brains were in the later, neurodegenerative phase (>65 yr old) (Additional file 1: Figure S5b). The same hpCGG slides were also examined for inclusion autofluorescence, which was not detected. Finally, very few ubiquitin-staining inclusions were found on the entire slide used to detect SUMO 2/3 immunofluorescence, consistent with the ~1.6% of cells with inclusions in frontal cortex for these premutation mice at 11 months of age [142], which was a much lower percentage of inclusion-bearing cells compared to SUMO 2/3 aggregate-bearing cells. This finding indicates that SUMO 2/3 aggregation precedes the formation of canonical ubiquitin-staining FXTAS inclusions.
The levels of SUMO 2/3 in nuclei from FXTAS brains exceed the levels found in several other common neurodegenerative disorders
To determine whether the elevated SUMO 2/3 in FXTAS is specific to this disorder or is representative of a generalized neurodegenerative phenomenon, a variety of other neurodegenerative brain samples were collected for nuclear isolation followed by western blot (Fig. 4 d, Additional file 1: Figure S6). No other disease consistently exhibited elevated SUMO 2/3 immunoreactivity to the same degree as FXTAS, but several other brain samples from patients exhibiting neurodegenerative disease did exhibit higher than normal levels of SUMO 2/3, including FTD, HD, and PSP. Although one patient diagnosed with AD did show an abnormally high SUMO 2/3 signal, this was not consistently recapitulated in other AD samples. Brains from patients diagnosed with PD, ALS, and FXS exhibited SUMO 2/3 levels similar to that of control brains. These results are preliminary, as more replicates were not possible due to insufficient sample quantity, but they indicate that the consistently high level of SUMO 2/3 seen in FXTAS samples is unique to this disorder; although, SUMO 2/3 may play a role in other inclusion and/or aggregate-bearing neurodegenerative diseases. Samples tested in the current study were nuclear lysates, so diseases bearing cytoplasmic-only inclusions, such as PD and ALS, may have aggregates that contain SUMO 2/3 in their cytoplasmic compartments that would not be captured here. The role of SUMO 2/3 in other neurodegenerative disorders warrants further study.
SUMO 2/3 immunoprecipitation proteomics reveal that SUMO 2/3 is highly conjugated to proteins involved with protein turnover, DNA damage/repair, and protection against oxidative stress/damage in FXTAS
To further assess the targets of SUMO 2/3 conjugation in FXTAS patients, we performed IP with a SUMO 2/3 antibody followed by LC-MS/MS to determine what population of proteins are modified by SUMO 2/3 in FXTAS patients compared to controls. IP methods were adapted from several sources [50, 52, 145, 155]. In brief, nuclei were isolated from frontal cortex tissue of two FXTAS cases and two controls. Nuclei were lysed and heated in an SDS/Triton/deoxycholate buffer, then diluted to minimize detergent interference for IP. Pulldown was performed using a SUMO 2/3 mouse IgG antibody and anti-mouse IgG magnetic beads. Western blot confirmed that the pulldown successfully captured the SUMO 2/3 smear (Additional file 1: Figure S7). LC-MS/MS of the IP eluted proteins confirmed the high levels of SUMO 2/3 pulldown in the FXTAS samples (Table 2). As with the MS analysis of the FXTAS inclusions, no one protein dominated in the IP, indicating that the elevated conjugated SUMO 2/3 present in FXTAS is not due to its conjugation with one particular protein.
Fold change values for the averaged FXTAS IP samples over the averaged age-matched control IP samples are shown. Proteins that were present in at least one FXTAS sample to at least 0.1% and showed at least 20% enrichment in both FXTAS samples over age matched control samples are displayed here. The high level of enrichment in SUMO 2 signal present in FXTAS samples confirms the success of the IP. Excluding SUMO 2, proteins that showed at least 3-fold enrichment in both FXTAS samples over age matched control samples are bolded. Numeric values presented outside parentheses are fold change of averaged FXTAS samples over averaged control samples. Values inside parentheses show the averaged abundance values as a percentage of the total protein composition identified for FXTAS samples over control samples.
Among the more abundant SUMO 2/3 targets identified by IP were proteins with chaperone/protein-folding functions, including αβ-crystallin (CRYAB) and peptidyl-prolyl cis-trans isomerase A (PPIA) [13, 48]; conjugating proteins known to be involved in protein turnover, including ubiquitin, SUMO 1, and SUMO 2/3 [79, 96, 102]; and prominently, the ubiquitin-binding shuttling protein p62 [26, 107, 143]. We also detected proteins known to be involved in non-homologous end joining (NHEJ), including poly [ADP-ribose] polymerase 1 (PARP1), X-ray repair cross-complementing protein 6 (XRCC6/Ku70), and X-ray repair cross-complementing protein 5 (XRCC5/Ku80) [22, 146]. Several proteins involved in cellular protection against oxidative stress, such as carbonyl reductase 1 (CBR1) [67, 121, 151] and mitochondrial isocitrate dehydrogenase (IDH2) [47, 61, 69], were also identified. Other than SUMO 2, only 4 proteins were enriched at least 3-fold in FXTAS IPs over control IPs: SUMO 1, SUMO 3, Ubiquitin, and p62.
FMRpolyG is a minor component in FXTAS inclusions and IP conjugates
It has been posited that FXTAS inclusions are largely made up of aggregates of FMRpolyG and interacting proteins [19, 125, 137]. However, endogenous FMRpolyG has never been identified in FXTAS patient samples through direct protein sequencing. Importantly, we have identified FMRpolyG in both sorted inclusion samples and FXTAS SUMO IP samples (Table 3). However, FMRpolyG constituted an extremely minor component, at 0.003% of the total protein molar complement in one inclusion sample and 0.04% in the other sample (~30–400 ppm molar abundance). Importantly, neither of the FXTAS nuclear lysate samples nor control brain contained detectable levels of FMRpolyG. Thus, FMRpolyG can only be detected with significant enrichment. Additionally, two proteins, LAP2β and TRA2A, which have been proposed to co-aggregate with FMRpolyG within inclusions [19, 125], were also found at very low levels. LAP2β was detected at 0.02% and 0.04% in inclusions while TRA2A was detected at 0.0001% in the inclusion samples. As a positive control for detection of FMRpolyG, we expressed an FMRpolyG GFP fusion construct in SK-N-MC neuroblastoma cells (Fig. 5 a). We identified three proteotypic peptides mapping to FMRpolyG-GFP that were not detected in null or GFP transfected cells (Fig. 5 b). Finally, FMRpolyG was also detected in both FXTAS IP samples, but was again at a low abundance (0.01%) and detected through just one hit of one tryptic peptide in each sample (Table 3).
a Vector pBR-CMV-5ATG-63-EGFP produces FMRpolyG containing 63 CGG repeats fused to EGFP under a strong CMV enhancer and promoter. As a positive control for FMRpolyG detection by LC-MS/MS, an expression plasmid was generated with a CMV promoter driven FMRpolyG-GFP fusion construct. The human FMR1 5′ untranslated region (UTR) and Exon 1 sequence, from Transcriptional Start Site I (TSS-I) to the first three base pairs of Exon 2, were placed upstream and in frame with EGFP to create a stable fusion protein. The translational start site of FMRpolyG was modified from the native ACG to the canonical ATG to further drive FMRpolyG expression. b LC-MS/MS analysis of FMRpolyG-GFP expressed in an SK-N-MC heterologous cell expression system identifies multiple proteotypic peptides. Transiently transfected SK-N-MC cells containing the pBR-CMV-5ATG-63-EGFP plasmid were SDS solubilized and whole cell protein lysates were trypsin digested and analyzed by LC-MS/MS. GFP signal was observed in FMRpolyG and GFP expressing cells, but not null transfection controls, indicating successful transfection of the constructs. Peptide sequence coverage of FMRpolyG-GFP (48%) is indicated in yellow with modified residues in green. LC-MS/MS identified three unique FMRpolyG-GFP peptides present only in FMRpolyG-GFP expressing cells that map to native FMRpolyG protein. A representative mass spectrum of the FMRpolyG specific peptide SPPLGGGLPALAGLK is provided with y/b ions labeled
FXTAS inclusions do not co-localize with the FMR1 gene
The observation that FXTAS inclusions are exclusively solitary and intranuclear has led to the hypothesis that inclusion formation may occur at the active FMR1 locus, perhaps forming due to the accumulation of DDR proteins around the damage-prone expanded repeat [32, 42, 43]. To test this, a DNA fluorescence in situ hybridization (FISH) composite probe set, comprised of 119 individual DNA oligonucleotides, was generated to target the human FMR1 locus. The FISH probe was generated using a modified PCR protocol to avoid the repeat-rich regions surrounding and including the FMR1 locus. Probe specificity and efficiency was tested on a variety of human cells, including metaphase and interphase fibroblasts cultured from skin biopsies, lymphocytes, and brain smears made from fresh frozen human frontal cortex tissue (Fig. 6 a). In fibroblasts, over one thousand nuclei from both female and male samples were scored for probe binding, and 97% of nuclei contained clear and specific FISH probe signal. In human brain nuclear smears, over 500 nuclei from one FXTAS patient were scored for FISH probe binding, inclusion presence, and localization of the FMR1 locus relative to inclusions. 92% of the scored nuclei contained clear and specific probe signal, and 8.7% of the scored nuclei contained nuclear inclusions (Fig. 6 b). Previous reports of inclusion load in FXTAS frontal cortical tissue range from 2–20% of neurons and astrocytes [38], and our quantification of inclusion load based on autofluorescence falls within this range. Of the inclusion- and probe-bearing nuclei, 95.2% of them clearly showed no-colocalization between the inclusion and the FMR1 locus, and the remaining 4.8% showed possible co-localization (Fig. 6 b). These data do not support the hypothesis that the FMR1 locus consistently co-localizes with FXTAS inclusions.
a DNA FISH probe generated for the FMR1 locus successfully tags the DNA locus in fibroblasts, lymphocytes, and brain nuclei. DNA FISH probe generated using PCR amplification to tag the FMR1 locus cleanly tags the FMR1 locus at high efficiency in all cell types examined. Metaphase plates generated using control patient-derived fibroblasts exhibit expected tagging patterns, with male samples exhibiting one tag located towards the end of the long arm of the X chromosome, and with female samples exhibiting two such tags. Scoring of over 1100 fibroblast cells in a grid-like fashion showed 97% of cells exhibited sensitive and specific binding. Patient-derived lymphocytes exhibited similar binding and efficiency. Brain nuclei exhibited slightly lowered binding efficiency at 92%, possibly due to degradation of cells due to pre and postmortem conditions. b The FMR1 locus does not preferentially co-localize with inclusions. Out of 563 FXTAS nuclei scored, 518 of the nuclei displayed sensitive and specific probe binding, representing an approximate FISH probe efficiency of 92% in brain tissue. Forty-five nuclei had either no probe signal or multiple probe signals. Forty-nine of the nuclei scored positive for inclusions, representing about 8.7% of the total nuclei present. Forty-two of inclusion-bearing nuclei also showed sensitive and specific probe binding. Forty of these nuclei showed negative colocalization between the FMR1 probe and inclusion, while 2 nuclei showed possible colocalization between the FMR1 probe and the inclusion. These data indicate that the FMR1 gene does not co-localize with FXTAS inclusions
Discussion
A principal outcome of the current study is that the protein complement of the FXTAS intranuclear inclusions is not dominated by a single, enriched protein. Rather, the inclusions comprise a large number of proteins, each present at only a few percent or less of the total proteins identified. The current observations both validate and expand upon an earlier, more limited study of FXTAS inclusions [58]; with a number of proteins identified previously now quantified (see: Additional file 1) as well as many additional protein components identified (e.g. SUMO 2/3, p62). Moreover, the current study has followed up the finding of SUMO2/3 enrichment by using immunoprecipitation to identify proteins to which these modifiers are ligated.
The strong enrichment for proteins involved in protein turnover as well as the overall heterogeneous population indicates that the inclusions are principally repositories of proteins destined for removal. There are several possibilities for why these proteins are aggregating. There is good evidence for the presence of RNA in the inclusions, and particularly FMR1 mRNA [135], which may serve as driver for aggregation. In support of the involvement of RNA, RNase treatment does affect the integrity of the inclusions (Fig. 2), and 36% of inclusion proteins are RNA binding proteins, which further supports the presence of RNA (Fig. 4). Although RNA sequencing of FXTAS inclusions would be an excellent way to ascertain what RNA species might be playing roles in inclusion formation, efforts to characterize the RNA within the inclusions have not been successful thus far due to extensive degradation of the RNA isolates. This could be due to either the strategy used in this study to isolate inclusions or due to intrinsically degraded RNAs within the inclusions.
In addition, SUMO 2 may play a role in aggregation. SUMO proteins have been well-studied in the context of neurodegenerative disease for the diverse roles that they can play as protein modifiers [4]. Although SUMO 2 was found to co-aggregate with polyQ-ATXN7, and mouse models showed accumulation of SUMO 2 in SCA7 patient brain [90], SUMO 2 had not previously been reported as a major inclusion protein. The intensity of the SUMO 2 signal in FXTAS compared to other neurodegenerative diseases suggests something specific in FXTAS which is not present or not as apparent in other inclusion disorders. Unlike the current study, almost all studies reporting a role for SUMO in other neurodegenerative diseases have found roles for SUMO 1 rather than SUMO 2/3 [29, 85, 132, 152]. SUMO 1 is the dominant SUMO species and the majority of target proteins are exclusively modified by SUMO 1 [149]. However, in response to cellular stress, including heat shock, oxidative stress, and DNA damage, SUMO 2/3 forms polySUMO chains, while SUMO 1 may only cap these chains [4, 140]. A study in HD found higher levels of high molecular weight SUMO 2/3 in insoluble fractions of HD affected striatum and suggested that this resulted from toxic mutant huntingtin protein inducing a cellular stress response [105]. Increased oxidative stress and DNA damage in FXTAS have been well reported [2, 117, 120, 131]. PolySUMOylation has been found to drive formation of biomolecular condensates through multivalent interactions [116], so it is plausible that the polySUMO chains found in FXTAS inclusions may be a driver of, or participate in the formation of inclusions. Since SUMO 2 aggregation appears to precede formation of canonical ubiquitin-containing inclusions, it is possible that SUMO 2 aggregation helps initiate inclusion formation.
Beyond polySUMOylation, other inclusion-enriched proteins point to a global increase in DNA damage. SUMO 2 and ubiquitin are both found at high levels in FXTAS inclusions modifying a large pool of proteins, and SUMO 2 IP was able to pull down large amounts of ubiquitin in FXTAS samples. Therefore, it is likely that mixed ubiquitin/SUMO chains are present. There have been reports that ubiquitin and SUMO 2 compete for the same modification sites on proteins, and may have opposing effects [75, 139], but SUMO 2/3 is also known to form mixed chains with ubiquitin that can act cooperatively to recruit DDR mediators to damaged sites or enhance protein degradation [4, 41]. Moreover, increased modification of proteins by SUMO-2/3 is a cytoprotective response against cell stress [86], which is clearly involved with FXTAS pathogenesis. p62 is also a known player in DDR response, and is known to be a cargo receptor that directs tagged proteins destined for degradation by the autophagosome and has previously been considered a major component of cytoplasmic inclusions [80, 154]. We speculate that its binding dynamics are not significantly different in the nucleus. Monomeric p62 is able to bind ubiquitinated substrates through its ubiquitin-associated domain [56], and it has been shown to actively participate in DNA damage repair, acting as a shuttle to transport modified proteins to the UPS or the autophagosome for removal [51, 143]. In addition, p62 regulation has been directly linked to calpain protease activity [20, 156], which may provide a link between the results presented here and Ca and calpain dysregulation seen previously in FXTAS [117].
Recently, we demonstrated that μ-calpain activity was chronically elevated in cortical and hippocampal tissues prepared from mice expressing FMR1 CGG expansions repeats in the high range (premutation mice), beginning as early as age 6 weeks and reaching >2-fold that of age matched WT mice by 6 months [117]. The increased μ-calpain activity is the likely consequence of chronically elevated cytoplasmic Ca2+ and oxidative stress within premutation neurons [117]. Elevated μ-calpain activity in premutation brain was also associated with a 2-fold higher p25/p35 ratio, dysregulation of Cdk5, and elevated P-Ser1981-ATM – all by 6 months. These findings were in accord with corresponding molecular outcome measures in premutation hippocampal neurons at 7 DIV, the latter outcomes all normalized by the ER-Ca2+ channel inhibitor dantrolene [117]. Neuronal μ-calpain activity has been implicated as a primary mechanism regulating p62, which acts as an autophagic receptor that recognizes misfolded protein aggregates targeted for lysosomal degradation [12, 33, 95]. Adaptive autophagy in response to conditions of oxidative stress can have protective or pathological influences on neurons. Basal autophagy is important for the turnover of organelles and proteins preventing accumulation neurotoxic aggregates associated with neurodegeneration [71]. However, autophagy is vulnerable to other stress signals that can disrupt adaptive functions, with abnormal Ca2+/calpain regulation providing a mechanistic link between autophagy and apoptosis [103, 150], suggesting that chronically elevated Ca2+ and calpain activation may drive an autophagic survival response into maladaptive autophagy that contributes to neuronal death in FXTAS. Considering the significantly elevated levels of p62 and SUMO2 in the nuclear compartment and inclusions of FXTAS brains, we posit involvement of maladaptive autophagy in the neurodegenerative phase of FXTAS. This may not be overly speculative as ATM-mediated DNA repair is a consequence of p62 accumulation and has been linked to neurodegeneration [63, 81, 141].
Based on the current data, we propose a model for inclusion formation (Fig. 7) wherein a cellular stress response from a global increase in ROS and mitochondrial dysfunction [100, 117, 120], perhaps exacerbated by ROS-induced DNA damage, results in increased load of damaged/oxidized proteins as well as proteins that are involved with DDR. Ubiquitin and SUMO 2/3 target these proteins for degradation. However, if the production of these damaged and/or tagged proteins exceeds the capacity of the proteasomal machinery within the nucleus, the proteins alone or in combination with various mRNA species (e.g., FMR1 CGG repeat mRNA) known to be present in the inclusions [135] may drive aggregation. Macroscopic aggregation would appear as an inclusion, which would not be possible for the UPS to degrade, and may even impair proteasomal function [80]. Autophagy is the normal cellular mechanism for aggregate degradation, and p62 binds the aggregation to ferry it to the autophagosome for removal [143], but beyond a certain point, the inclusion becomes too large to exit the nucleus, resulting in a chronic nuclear accumulation. The UPS system has been shown to exhibit decreased function with aging, and there has been some evidence of UPS impairment in other neurodegenerative diseases, which may explain why FXTAS symptoms and inclusion formation are most apparent at a later age [9, 66, 84, 99]. In addition, inhibition of the UPS system has been shown to generate inclusion-like structures, which further supports the connection between inclusion formation and UPS impairment [76, 77, 110].
Diagrammatic representation of hypothesized FXTAS inclusion formation. Within FXTAS brain nuclei, proteins (yellow hexagons) destined for removal are tagged with ubiquitin and SUMO 2/3 chains, which are bound by the UPS for degradation. Polyubiquitinated and polySUMOylated proteins may form small aggregates with each other and/or RNA, at which point p62 will shuttle the aggregate out of the nucleus to an autophagosome for removal. Over time, as the FXTAS patient experiences higher levels of oxidative stress and DNA damage and decreased functioning of the UPS due to aging or injury, the levels of damaged/oxidized proteins and DNA damage mediators requiring removal increase. Paired with decreased UPS functionality, these proteins get tagged for removal but build up in the nucleus, aggregating with other proteins and RNA. p62 may attempt to shuttle the aggregate out to the autophagosome, but if the accumulation becomes too large, p62 has no way of shuttling the mass out of the nucleus in post-mitotic cells, resulting in an inclusion
One interesting result of the current study was the finding of extremely low levels of FMRpolyG either within inclusions or in FXTAS nuclei. It has been proposed that FMRpolyG, in combination with proteins such as LAP2β and TRA2A, aggregate to form FXTAS inclusions [15, 19, 118, 125, 137]. Our study is the first to identify endogenous FMRpolyG in FXTAS patient samples by direct protein sequencing. However, although the current study did detect minute quantities of FMRpolyG in inclusions and in SUMO 2/3 IP-enriched samples, the levels render it unlikely that FMRpolyG is a main driver or material participant in the formation of FXTAS inclusions. Moreover, FMRpolyG was undetectable in total nuclear samples, indicating that it exists at extremely low endogenous levels. Although LAP2β and TRA2A were also detected in inclusions, they were also found at very low levels (Additional file 1). LAP2 β is estimated to be present at about the 0.02-0.04% level, whereas TRA2A is present only at ~0.0002% (2 ppm) and is actually about 10- to 15-fold lower in the inclusions than in the surrounding nuclear milieu. Therefore, although LAP2β AND TRA2A have been observed by others to co-localize with FMRpolyG in FXTAS inclusions [19, 125], it is unlikely that these proteins substantively contribute to inclusion formation.
Part of the difficulty in assessing the involvement of potential inclusion proteins such as FMRpolyG, LAP2 β, and TRA2A by immunocytochemistry is that the method, though great for localization of proteins, is not a sound method for quantification, since antibody staining efficiency and methodology can have large effects on the intensity of staining. LAP2β and TRA2A are cases in point, as previously published immunofluorescence studies have demonstrated the presence of these proteins in inclusions, yet the actual levels of these proteins in inclusions are extremely low. As another example, we performed immunofluorescence on FXTAS inclusions using previously published antibodies against FMRpolyG [15], and we found highly variable staining patterns in inclusions, varying from very faint, circumscribed, and very bright, all on the same slide (Additional file 1: Figure S8a). Thus, although the RAN translation pathogenic model cannot be ruled out by the current results, the current findings underscore the need for more studies that involve endogenous expression rather than in vitro, induced, or high-expression models. Finally, the current study also shows that co-localization between the FMR1 gene and FXTAS inclusions does not consistently occur, which rules out DDR as a response to damage at the FMR1 locus as the initiator of inclusion formation.
The current study used a novel method to purify endogenous FXTAS inclusions from human brain whereby the intrinsic autofluorescence of the inclusions was used as a means for their separation, via preparative FACS, from other nuclear particles and organelles (e.g., nucleoli). The vast majority of studies of the protein composition of inclusions and other nuclear or cytoplasmic aggregates in neurodegenerative disorders, including our own [6, 58], have relied on immunocytochemistry and other staining methods [1, 3, 31, 65, 73, 98, 110]. Such methods depend not only on the uniform presence and accessibility of the specific proteins being probed, but also on the properties of antibody sensitivity and specificity. Moreover, many studies have utilized in vitro induction of inclusion-like bodies, which may not fully recapitulate endogenous conditions [77, 91, 97, 133, 153, 157]. Among the few studies that have purified endogenous inclusions from human patients for proteomics, methods for purification have relied on immunolabeling or sequential extraction by detergents or chaotropes for which it was assumed that the most insoluble fraction was purified inclusion [37, 62, 93, 101, 112, 128, 144]. These methods introduce bias if antibodies were used, and insoluble fractions may or may not contain a truly pure population of inclusions. The method used in this study avoids these areas of bias by utilizing autofluorescent properties of FXTAS inclusions for FACS. As a concrete example of this distinction, the autofluorescent inclusions are quite distinct from the other major intranuclear organelle, the nucleolus, which is completely non-autofluorescent (Additional file 1: Figure S8b).
The discovery of autofluorescence in FXTAS inclusions allows us to more specifically identify and isolate inclusions, but also calls for more stringent guidelines when examining inclusions by immunofluorescence. This finding does not necessarily nullify past immunofluorescence results on inclusions, but such results should now be reassessed using immunofluorescence strategies that take autofluorescence into account. Such strategies are already in use in the field of NIID, which noted autofluorescent inclusions at certain wavelengths [70, 110]. More detailed recommendations will be published in a separate technical paper. However, in brief, FXTAS inclusion autofluorescence interferes most strongly in the green, yellow, and orange spectrum. Since the blue range of the spectrum is usually reserved for DAPI staining, efforts should be made whenever possible to perform inclusion immunofluorescence in the far-red spectrum. There are still low levels of autofluorescence in that spectral region. Therefore, slides with secondary-only antibody staining should be imaged using the same settings as those with primary antibody staining of inclusions and provided for comparison. If it is necessary to perform immunofluorescence at a wavelength where autofluorescence is substantial, the antibody used must first be verified to have sufficient sensitivity and specificity to overcome autofluorescence.
The use of an autofluorescence-based method of purifying inclusions is novel; therefore, it is important to verify that what is collected from FACS is a substantially pure population of inclusions. In the current instance, microscopy of the sorted inclusions verified that their autofluorescence profile was the same as that of inclusions in situ (Fig. 1 c). Moreover, western blot and immunofluorescence analyses verified that two of the newly-identified inclusion proteins that were most enriched by MS are increased both in FXTAS nuclei and in FXTAS inclusions (Fig. 6, Additional file 1: Figure S7). Interestingly, several of the proteins found to be enriched in FXTAS inclusions correspond to proteins found in the inclusions of other neurodegenerative disorders. Two of the FXTAS inclusion proteins with greatest enrichment – ubiquitin and p62 – have previously been well-documented. Ubiquitin is a component in some forms of FTD, HD, some forms of spinocerebellar ataxia (SCA), PD, ALS, dentatorubral-pallidoluysian atrophy (DRPLA), and NIID [11, 28, 49, 70, 72, 87, 123]. p62 has also been found in other inclusion disorders, including FTD, PD, and AD [49, 88, 154]. A third protein enriched in FXTAS inclusions, MLF2, has previously been found to co-aggregate with p62 and poly (gly-ala) in mice, and it alleviated poly-Q associated toxicity in Drosophila and rat models [36, 124]. The presence of MBP enrichment in FXTAS inclusions is not well understood, as MBP is traditionally known to be a cytoplasmic oligodendrocyte protein associated with myelination. However, variant forms of it have been found to inhabit oligodendrocyte nuclei [108] and to play unrelated functional roles in astrocytic and neuronal nuclei [74]. Regardless, it has previously been observed as a FXTAS inclusion protein [58] and further investigation into its role is warranted. In addition, the functional families of proteins found enriched in inclusions are similar to what has been seen in other types of inclusions. RNA binding proteins, and especially hnRNPs, have previously been detected in other types of inclusion disorders, as well as chaperones, particularly heat shock proteins [21, 24, 34, 65, 92, 111]. The presence of proteasomal proteins in FXTAS inclusions is not surprising as a connection between the UPS and inclusions has long been suspected.
Aside from comparison to inclusion composition in other disorders, the FXTAS inclusion composition was also compared to that of lipofuscin to rule out the possibility that lipofuscin was a significant component of sorted FXTAS inclusions. Lipofuscins are intracytoplasmic, perinuclear pigment granules that are composed of the residue of lysosomal degradation of aggregated/damaged proteins. It accumulates in normal aging human brain, and autofluoresces throughout a broad range of wavelengths. The main difference in inclusion and lipofuscin autofluorescence is in the far-red wavelengths, where lipofuscin autofluorescence is significantly more intense than inclusion autofluorescence (Additional file 1: Figure S1). It is unlikely that sorted inclusions contain a significant component of lipofuscin for the following reasons: (i) the nuclear isolation step should have largely eliminated extranuclear cell debris, which would include the cytoplasmic lipofuscin; (ii) the FACS sorted fraction from the nuclear isolates was only present in the FXTAS cases, not in controls, whereas lipofuscin is present in controls as well as FXTAS cases; (iii) the sorted inclusions were additionally viewed by microscopy to ensure that a minimal proportion of sorted particles were brightly fluorescent in the far-red wavelength (Fig. 1d); and (iv) we compared lipofuscin protein components as identified by MS in a previous publication to the composition of the FXTAS inclusions determined in the current study [106]. Out of 49 identified major lipofuscin proteins, 7 were not found in our dataset, 7 were found at extremely low levels (<0.005%), and almost all the proteins that were found in both datasets were not enriched in FXTAS inclusions over total nuclear samples.
The current investigation has some limitations that bear consideration. First, although the purification process for inclusions is robust and more specific than other inclusion isolation processes in the field, we estimate that as much as 10–20% of noninclusion material may remain in the final preparations. This could allow some proteins that are not within the inclusion to be detected by MS, although the comparison to accompanying nuclear samples – establishing levels of enrichment – should help to mitigate this problem. Second, due to the need for long sorting times and amounts of brain tissue required to isolate sufficient numbers of inclusions for MS, only two FXTAS patients were analyzed. Third, a limitation of the MS analysis is that the quantification and percentages of proteins composing a population is limited by the population of proteins detected. Therefore, if there are undetectable proteins due to insufficient solubility, lack of available tryptic cut sites, or unforeseen pos-translational modifications, these would not be represented in the protein population. Regarding FMRpolyG, it has been reported that solubilization may be challenging. However, we used mild heating in a strong detergent buffer, the same strength as that used in a previous western blot of FMRpolyG [137], as well as formic acid; we did not observe any precipitate formation. Previously, only two western blots for FMRpolyG on native human brain tissue have been published [125, 137], and both utilized thermal denaturation by boiling the tissue samples. It may be that solubilization problems arise due to heat-dependent denaturation/aggregation, as has been established previously [8, 40, 122, 138, 148].
Conclusions
We have performed a detailed analysis of the protein composition of isolated FXTAS inclusions using their distinct autofluorescent properties as a means for preparative FACS-based purification. Although the current study has helped to refine our understanding of the role played by inclusions in FXTAS pathogenesis, further investigation into the role of SUMO2/3 in FXTAS pathogenesis is warranted, as well as a re-investigation of the roles of FMRpolyG in this disorder. Finally, we wish to point out that models proposed for mRNA sequestration remain viable [60, 115, 126, 127, 130], and these models/mechanisms should be investigated further.
Availability of data and materials
The proteomics datasets generated and analyzed in the current study are available in the MassIVE repository (massive.ucsd.edu) under ID MSV000084199.
Abbreviations
- AD:
-
Alzheimer’s disease
- ALS:
-
amyotrophic lateral sclerosis
- BSA:
-
bovine serum albumin
- CBR1:
-
carbonyl reductase 1
- CERAD:
-
Consortium to Establish a Registry for Alzheimer’s Disease
- CRYAB:
-
αβ-crystallin
- DDA:
-
data-dependent acquisition
- DDR:
-
DNA damage response
- DM:
-
myotonic dystrophy
- DRPLA:
-
dentatorubral-pallidoluysian atrophy
- FACS:
-
fluorescence activated cell sorting
- FISH:
-
fluorescence in situ hybridization
- FTD:
-
frontotemporal dementia
- FXS:
-
fragile X syndrome
- FXTAS:
-
Fragile X-associated tremor/ataxia syndrome
- HCD:
-
high energy collisional dissociation
- HD:
-
Huntington’s Disease
- hnRNP:
-
heterogeneous nuclear ribonucleoprotein
- hpCGG:
-
high-premutation CGG
- IDH2:
-
isocitrate dehydrogenase
- IP:
-
immunoprecipitation
- LAP2β:
-
lamina-associated polypeptide 2 beta
- LC-MS/MS:
-
liquid chromatography/tandem mass spectrometry
- MBP:
-
myelin basic protein
- MLF2:
-
myeloid leukemia factor 2
- MS:
-
mass spectrometry
- NHEJ:
-
non-homologous end joining
- NIH:
-
National Institutes of Health
- NIID:
-
neuronal intranuclear inclusion disease
- PARP1:
-
poly [ADP-ribose] polymerase 1
- PD:
-
Parkinson’s disease
- PPIA:
-
peptidyl-prolyl cis-trans isomerase A
- PSP:
-
progressive supranuclear palsy
- RAN:
-
repeat-associated non-ATG
- SCA:
-
spinocerebellar ataxia
- SQSTM1:
-
sequestosome1
- STED:
-
stimulated emission depletion
- SUMO 2:
-
small ubiquitin-related modifier 2
- TBS:
-
tris buffered saline
- TEAB:
-
triethylammonium bicarbonate
- TRA2A:
-
transformer-2 protein homolog alpha
- UCD:
-
University of California, Davis
- WT:
-
wildtype
- XRCC5:
-
x-ray repair cross-complementing protein 5
- XRCC6:
-
x-ray repair cross-complementing protein 6
References
Adachi H, Waza M, Tokui K, Katsuno M, Minamiyama M, Tanaka F, Doyu M, Sobue G (2007) CHIP overexpression reduces mutant androgen receptor protein and ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model. J Neurosci 27:5115–5126. https://doi.org/10.1523/JNEUROSCI.1242-07.2007
Alvarez-Mora MI, Rodriguez-Revenga L, Madrigal I, Guitart-Mampel M, Garrabou G, Mila M (2017) Impaired Mitochondrial Function and Dynamics in the Pathogenesis of FXTAS. Mol Neurobiol 54:6896–6902. https://doi.org/10.1007/s12035-016-0194-7
Amador-Ortiz C, Lin WL, Ahmed Z, Personett D, Davies P, Duara R, Graff-Radford NR, Hutton ML, Dickson DW (2007) TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer's disease. Ann Neurol 61:435–445. https://doi.org/10.1002/ana.21154
Anderson DB, Wilkinson KA, Henley JM (2009) Protein SUMOylation in neuropathological conditions. Drug News Perspect 22:255–265. https://doi.org/10.1358/dnp.2009.22.5.1378636
Ariza J, Rogers H, Monterrubio A, Reyes-Miranda A, Hagerman PJ, Martinez-Cerdeno V (2016) A Majority of FXTAS Cases Present with Intranuclear Inclusions Within Purkinje Cells. Cerebellum 15:546–551. https://doi.org/10.1007/s12311-016-0776-y
Arocena DG, Iwahashi CK, Won N, Beilina A, Ludwig AL, Tassone F, Schwartz PH, Hagerman PJ (2005) Induction of inclusion formation and disruption of lamin A/C structure by premutation CGG-repeat RNA in human cultured neural cells. Hum Mol Genet 14:3661–3671. https://doi.org/10.1093/hmg/ddi394
Aziz A, Fronczek R, Maat-Schieman M, Unmehopa U, Roelandse F, Overeem S, van Duinen S, Lammers GJ, Swaab D, Roos R (2008) Hypocretin and melanin-concentrating hormone in patients with Huntington disease. Brain Pathol 18:474–483. https://doi.org/10.1111/j.1750-3639.2008.00135.x
Benjwal S, Verma S, Rohm KH, Gursky O (2006) Monitoring protein aggregation during thermal unfolding in circular dichroism experiments. Protein Sci 15:635–639. https://doi.org/10.1110/ps.051917406
Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR (2007) Global changes to the ubiquitin system in Huntington's disease. Nature 448:704–708. https://doi.org/10.1038/nature06022
Berman RF, Buijsen RA, Usdin K, Pintado E, Kooy F, Pretto D, Pessah IN, Nelson DL, Zalewski Z, Charlet-Bergeurand N, Willemsen R, Hukema RK (2014) Mouse models of the fragile X premutation and fragile X-associated tremor/ataxia syndrome. J Neurodev Disord 6:25. https://doi.org/10.1186/1866-1955-6-25
Bigio EH, Johnson NA, Rademaker AW, Fung BB, Mesulam MM, Siddique N, Dellefave L, Caliendo J, Freeman S, Siddique T (2004) Neuronal ubiquitinated intranuclear inclusions in familial and non-familial frontotemporal dementia of the motor neuron disease type associated with amyotrophic lateral sclerosis. J Neuropathol Exp Neurol 63:801–811. https://doi.org/10.1093/jnen/63.8.801
Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614. https://doi.org/10.1083/jcb.200507002
Blair LJ, Baker JD, Sabbagh JJ, Dickey CA (2015) The emerging role of peptidyl-prolyl isomerase chaperones in tau oligomerization, amyloid processing, and Alzheimer's disease. J Neurochem 133:1–13. https://doi.org/10.1111/jnc.13033
Botta-Orfila T, Tartaglia GG, Michalon A (2016) Molecular Pathophysiology of Fragile X-Associated Tremor/Ataxia Syndrome and Perspectives for Drug Development. Cerebellum 15:599–610. https://doi.org/10.1007/s12311-016-0800-2
Buijsen RA, Sellier C, Severijnen LA, Oulad-Abdelghani M, Verhagen RF, Berman RF, Charlet-Berguerand N, Willemsen R, Hukema RK (2014) FMRpolyG-positive inclusions in CNS and non-CNS organs of a fragile X premutation carrier with fragile X-associated tremor/ataxia syndrome. Acta Neuropathol Commun 2:162. https://doi.org/10.1186/s40478-014-0162-2
Cao Z, Hulsizer S, Tassone F, Tang HT, Hagerman RJ, Rogawski MA, Hagerman PJ, Pessah IN (2012) Clustered burst firing in FMR1 premutation hippocampal neurons: amelioration with allopregnanolone. Hum Mol Genet 21:2923–2935. https://doi.org/10.1093/hmg/dds118
Chai Y, Wu L, Griffin JD, Paulson HL (2001) The role of protein composition in specifying nuclear inclusion formation in polyglutamine disease. J Biol Chem 276:44889–44897. https://doi.org/10.1074/jbc.M106575200
Chen Y, Tassone F, Berman RF, Hagerman PJ, Hagerman RJ, Willemsen R, Pessah IN (2010) Murine hippocampal neurons expressing Fmr1 gene premutations show early developmental deficits and late degeneration. Hum Mol Genet 19:196–208. https://doi.org/10.1093/hmg/ddp479
Cid-Samper F, Gelabert-Baldrich M, Lang B, Lorenzo-Gotor N, Ponti RD, Severijnen L, Bolognesi B, Gelpi E, Hukema RK, Botta-Orfila T, Tartaglia GG (2018) An Integrative Study of Protein-RNA Condensates Identifies Scaffolding RNAs and Reveals Players in Fragile X-Associated Tremor/Ataxia Syndrome. Cell Rep 25(3422-3434):e3427. https://doi.org/10.1016/j.celrep.2018.11.076
Colunga A, Bollino D, Schech A, Aurelian L (2014) Calpain-dependent clearance of the autophagy protein p62/SQSTM1 is a contributor to DeltaPK oncolytic activity in melanoma. Gene Ther 21:371–378. https://doi.org/10.1038/gt.2014.6
Cooper-Knock J, Walsh MJ, Higginbottom A, Robin Highley J, Dickman MJ, Edbauer D, Ince PG, Wharton SB, Wilson SA, Kirby J, Hautbergue GM, Shaw PJ (2014) Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain 137:2040–2051. https://doi.org/10.1093/brain/awu120
Couto CA, Wang HY, Green JC, Kiely R, Siddaway R, Borer C, Pears CJ, Lakin ND (2011) PARP regulates nonhomologous end joining through retention of Ku at double-strand breaks. J Cell Biol 194:367–375. https://doi.org/10.1083/jcb.201012132
Cunningham CL, Martinez Cerdeno V, Navarro Porras E, Prakash AN, Angelastro JM, Willemsen R, Hagerman PJ, Pessah IN, Berman RF, Noctor SC (2011) Premutation CGG-repeat expansion of the Fmr1 gene impairs mouse neocortical development. Hum Mol Genet 20:64–79. https://doi.org/10.1093/hmg/ddq432
Dammer EB, Fallini C, Gozal YM, Duong DM, Rossoll W, Xu P, Lah JJ, Levey AI, Peng J, Bassell GJ, Seyfried NT (2012) Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination. PLoS One 7:e38658. https://doi.org/10.1371/journal.pone.0038658
de Haro M, Al-Ramahi I, De Gouyon B, Ukani L, Rosa A, Faustino NA, Ashizawa T, Cooper TA, Botas J (2006) MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum Mol Genet 15:2138–2145. https://doi.org/10.1093/hmg/ddl137
Demishtein A, Fraiberg M, Berko D, Tirosh B, Elazar Z, Navon A (2017) SQSTM1/p62-mediated autophagy compensates for loss of proteasome polyubiquitin recruiting capacity. Autophagy 13:1697–1708. https://doi.org/10.1080/15548627.2017.1356549
Di Guardo G (2015) Lipofuscin, lipofuscin-like pigments and autofluorescence. Eur J Histochem 59:2485. https://doi.org/10.4081/ejh.2015.2485
DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277:1990–1993. https://doi.org/10.1126/science.277.5334.1990
Dorval V, Fraser PE (2006) Small ubiquitin-like modifier (SUMO) modification of natively unfolded proteins tau and alpha-synuclein. J Biol Chem 281:9919–9924. https://doi.org/10.1074/jbc.M510127200
Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD (2002) Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet 11:805–814. https://doi.org/10.1093/hmg/11.7.805
Fujigasaki H, Uchihara T, Koyano S, Iwabuchi K, Yagishita S, Makifuchi T, Nakamura A, Ishida K, Toru S, Hirai S, Ishikawa K, Tanabe T, Mizusawa H (2000) Ataxin-3 is translocated into the nucleus for the formation of intranuclear inclusions in normal and Machado-Joseph disease brains. Exp Neurol 165:248–256. https://doi.org/10.1006/exnr.2000.7479
Garcia-Arocena D, Hagerman PJ (2010) Advances in understanding the molecular basis of FXTAS. Hum Mol Genet 19:R83–R89. https://doi.org/10.1093/hmg/ddq166
Geronimo-Olvera C, Montiel T, Rincon-Heredia R, Castro-Obregon S, Massieu L (2017) Autophagy fails to prevent glucose deprivation/glucose reintroduction-induced neuronal death due to calpain-mediated lysosomal dysfunction in cortical neurons. Cell Death Dis 8:e2911. https://doi.org/10.1038/cddis.2017.299
Gittings LM, Foti SC, Benson BC, Gami-Patel P, Isaacs AM, Lashley T (2019) Heterogeneous nuclear ribonucleoproteins R and Q accumulate in pathological inclusions in FTLD-FUS. Acta Neuropathol Commun 7:18. https://doi.org/10.1186/s40478-019-0673-y
Glineburg MR, Todd PK, Charlet-Berguerand N, Sellier C (2018) Repeat-associated non-AUG (RAN) translation and other molecular mechanisms in Fragile X Tremor Ataxia Syndrome. Brain Res 1693:43–54. https://doi.org/10.1016/j.brainres.2018.02.006
Gobert V, Haenlin M, Waltzer L (2012) Myeloid leukemia factor: a return ticket from human leukemia to fly hematopoiesis. Transcription 3:250–254. https://doi.org/10.4161/trns.21490
Gozal YM, Dammer EB, Duong DM, Cheng D, Gearing M, Rees HD, Peng J, Lah JJ, Levey AI (2011) Proteomic analysis of hippocampal dentate granule cells in frontotemporal lobar degeneration: application of laser capture technology. Front Neurol 2:24. https://doi.org/10.3389/fneur.2011.00024
Greco CM, Berman RF, Martin RM, Tassone F, Schwartz PH, Chang A, Trapp BD, Iwahashi C, Brunberg J, Grigsby J, Hessl D, Becker EJ, Papazian J, Leehey MA, Hagerman RJ, Hagerman PJ (2006) Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS). Brain 129:243–255. https://doi.org/10.1093/brain/awh683
Greco CM, Hagerman RJ, Tassone F, Chudley AE, Del Bigio MR, Jacquemont S, Leehey M, Hagerman PJ (2002) Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain 125:1760–1771. https://doi.org/10.1093/brain/awf184
Gursky O, Aleshkov S (2000) Temperature-dependent beta-sheet formation in beta-amyloid Abeta (1-40) peptide in water: uncoupling beta-structure folding from aggregation. Biochim Biophys Acta 1476:93–102. https://doi.org/10.1016/s0167-4838(99)00228-9
Guzzo CM, Matunis MJ (2013) Expanding SUMO and ubiquitin-mediated signaling through hybrid SUMO-ubiquitin chains and their receptors. Cell Cycle 12:1015–1017. https://doi.org/10.4161/cc.24332
Hagerman P (2013) Fragile X-associated tremor/ataxia syndrome (FXTAS): pathology and mechanisms. Acta Neuropathol 126:1–19. https://doi.org/10.1007/s00401-013-1138-1
Hagerman PJ, Hagerman RJ (2015) Fragile X-associated tremor/ataxia syndrome. Ann N Y Acad Sci 1338:58–70. https://doi.org/10.1111/nyas.12693
Hagerman RJ, Hagerman P (2016) Fragile X-associated tremor/ataxia syndrome - features, mechanisms and management. Nat Rev Neurol 12:403–412. https://doi.org/10.1038/nrneurol.2016.82
Hall DA, Berry-Kravis E (2018) Fragile X syndrome and fragile X-associated tremor ataxia syndrome. Handbook of clinical neurology 147:377–391. https://doi.org/10.1016/b978-0-444-63233-3.00025-7
Hall DA, Robertson E, Shelton AL, Losh MC, Mila M, Moreno EG, Gomez-Anson B, Martinez-Cerdeno V, Grigsby J, Lozano R, Hagerman R, Maria LS, Berry-Kravis E, O'Keefe JA (2016) Update on the Clinical, Radiographic, and Neurobehavioral Manifestations in FXTAS and FMR1 Premutation Carriers. Cerebellum 15:578–586. https://doi.org/10.1007/s12311-016-0799-4
Han SJ, Jang HS, Noh MR, Kim J, Kong MJ, Kim JI, Park JW, Park KM (2017) Mitochondrial NADP(+)-Dependent Isocitrate Dehydrogenase Deficiency Exacerbates Mitochondrial and Cell Damage after Kidney Ischemia-Reperfusion Injury. J Am Soc Nephrol 28:1200–1215. https://doi.org/10.1681/ASN.2016030349
Haslbeck M, Vierling E (2015) A first line of stress defense: small heat shock proteins and their function in protein homeostasis. J Mol Biol 427:1537–1548. https://doi.org/10.1016/j.jmb.2015.02.002
Henderson MX, Chung CH, Riddle DM, Zhang B, Gathagan RJ, Seeholzer SH, Trojanowski JQ, Lee VMY (2017) Unbiased Proteomics of Early Lewy Body Formation Model Implicates Active Microtubule Affinity-Regulating Kinases (MARKs) in Synucleinopathies. J Neurosci 37:5870–5884. https://doi.org/10.1523/JNEUROSCI.2705-16.2017
Hendriks IA, Lyon D, Su D, Skotte NH, Daniel JA, Jensen LJ, Nielsen ML (2018) Site-specific characterization of endogenous SUMOylation across species and organs. Nat Commun 9:2456. https://doi.org/10.1038/s41467-018-04957-4
Hewitt G, Carroll B, Sarallah R, Correia-Melo C, Ogrodnik M, Nelson G, Otten EG, Manni D, Antrobus R, Morgan BA, von Zglinicki T, Jurk D, Seluanov A, Gorbunova V, Johansen T, Passos JF, Korolchuk VI (2016) SQSTM1/p62 mediates crosstalk between autophagy and the UPS in DNA repair. Autophagy 12:1917–1930. https://doi.org/10.1080/15548627.2016.1210368
Hilgarth RS, Sarge KD (2005) Detection of sumoylated proteins. Methods Mol Biol 301:329–338. https://doi.org/10.1385/1-59259-895-1:329
Hoem G, Bowitz Larsen K, Overvatn A, Brech A, Lamark T, Sjottem E, Johansen T (2019) The FMRpolyGlycine Protein Mediates Aggregate Formation and Toxicity Independent of the CGG mRNA Hairpin in a Cellular Model for FXTAS. Front Genet 10:249. https://doi.org/10.3389/fgene.2019.00249
Hoem G, Raske CR, Garcia-Arocena D, Tassone F, Sanchez E, Ludwig AL, Iwahashi CK, Kumar M, Yang JE, Hagerman PJ (2011) CGG-repeat length threshold for FMR1 RNA pathogenesis in a cellular model for FXTAS. Hum Mol Genet 20:2161–2170. https://doi.org/10.1093/hmg/ddr101
Hunsaker MR, Greco CM, Tassone F, Berman RF, Willemsen R, Hagerman RJ, Hagerman PJ (2011) Rare intranuclear inclusions in the brains of 3 older adult males with fragile x syndrome: implications for the spectrum of fragile x-associated disorders. J Neuropathol Exp Neurol 70:462–469. https://doi.org/10.1097/NEN.0b013e31821d3194
Isogai S, Morimoto D, Arita K, Unzai S, Tenno T, Hasegawa J, Sou YS, Komatsu M, Tanaka K, Shirakawa M, Tochio H (2011) Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J Biol Chem 286:31864–31874. https://doi.org/10.1074/jbc.M111.259630
Iwahashi C, Hagerman PJ (2008) Isolation of pathology-associated intranuclear inclusions. Methods Mol Biol 463:181–190. https://doi.org/10.1007/978-1-59745-406-3_13
Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, Babineau B, Lebrilla CB, Hagerman RJ, Hagerman PJ (2006) Protein composition of the intranuclear inclusions of FXTAS. Brain 129:256–271. https://doi.org/10.1093/brain/awh650
Iwatsubo T (2003) Aggregation of alpha-synuclein in the pathogenesis of Parkinson's disease. J Neurol 250(Suppl 3):III11–III14. https://doi.org/10.1007/s00415-003-1303-x
Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, Liu H, Feng Y, Warren ST (2007) Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron 55:556–564. https://doi.org/10.1016/j.neuron.2007.07.020
Jo SH, Son MK, Koh HJ, Lee SM, Song IH, Kim YO, Lee YS, Jeong KS, Kim WB, Park JW, Song BJ, Huh TL (2001) Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J Biol Chem 276:16168–16176. https://doi.org/10.1074/jbc.M010120200
Kametani F, Obi T, Shishido T, Akatsu H, Murayama S, Saito Y, Yoshida M, Hasegawa M (2016) Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Sci Rep 6:23281. https://doi.org/10.1038/srep23281
Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, Lu T, Yankner BA, Campisi J, Elledge SJ (2015) The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 349:aaa5612. https://doi.org/10.1126/science.aaa5612
Kaplan ES, Cao Z, Hulsizer S, Tassone F, Berman RF, Hagerman PJ, Pessah IN (2012) Early mitochondrial abnormalities in hippocampal neurons cultured from Fmr1 pre-mutation mouse model. J Neurochem 123:613–621. https://doi.org/10.1111/j.1471-4159.2012.07936.x
Kawamoto Y, Akiguchi I, Shirakashi Y, Honjo Y, Tomimoto H, Takahashi R, Budka H (2007) Accumulation of Hsc70 and Hsp70 in glial cytoplasmic inclusions in patients with multiple system atrophy. Brain Res 1136:219–227. https://doi.org/10.1016/j.brainres.2006.12.049
Keller JN, Hanni KB, Markesbery WR (2000) Impaired proteasome function in Alzheimer's disease. J Neurochem 75:436–439. https://doi.org/10.1046/j.1471-4159.2000.0750436.x
Kelner MJ, Estes L, Rutherford M, Uglik SF, Peitzke JA (1997) Heterologous expression of carbonyl reductase: demonstration of prostaglandin 9-ketoreductase activity and paraquat resistance. Life Sci 61:2317–2322. https://doi.org/10.1016/s0024-3205(97)00935-1
Kenneson A, Zhang F, Hagedorn CH, Warren ST (2001) Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum Mol Genet 10:1449–1454. https://doi.org/10.1093/hmg/10.14.1449
Kim SJ, Yune TY, Han CT, Kim YC, Oh YJ, Markelonis GJ, Oh TH (2007) Mitochondrial isocitrate dehydrogenase protects human neuroblastoma SH-SY5Y cells against oxidative stress. J Neurosci Res 85:139–152. https://doi.org/10.1002/jnr.21106
Kimber TE, Blumbergs PC, Rice JP, Hallpike JF, Edis R, Thompson PD, Suthers G (1998) Familial neuronal intranuclear inclusion disease with ubiquitin positive inclusions. J Neurol Sci 160:33–40. https://doi.org/10.1016/s0022-510x(98)00169-5
Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884. https://doi.org/10.1038/nature04723
Koyano S, Uchihara T, Fujigasaki H, Nakamura A, Yagishita S, Iwabuchi K (1999) Neuronal intranuclear inclusions in spinocerebellar ataxia type 2: triple-labeling immunofluorescent study. Neurosci Lett 273:117–120. https://doi.org/10.1016/s0304-3940(99)00656-4
Kurosawa M, Matsumoto G, Kino Y, Okuno M, Kurosawa-Yamada M, Washizu C, Taniguchi H, Nakaso K, Yanagawa T, Warabi E, Shimogori T, Sakurai T, Hattori N, Nukina N (2015) Depletion of p62 reduces nuclear inclusions and paradoxically ameliorates disease phenotypes in Huntington's model mice. Hum Mol Genet 24:1092–1105. https://doi.org/10.1093/hmg/ddu522
Landry CF, Ellison JA, Pribyl TM, Campagnoni C, Kampf K, Campagnoni AT (1996) Myelin basic protein gene expression in neurons: developmental and regional changes in protein targeting within neuronal nuclei, cell bodies, and processes. J Neurosci 16:2452–2462
Li S, Wang M, Qu X, Xu Z, Yang Y, Su Q, Wu H (2016) SUMOylation of PES1 upregulates its stability and function via inhibiting its ubiquitination. Oncotarget 7:50522–50534. https://doi.org/10.18632/oncotarget.10494
Li X, Zhang Y, Hu Y, Chang M, Liu T, Wang D, Zhang Y, Zhang L, Hu L (2008) Chaperone proteins identified from synthetic proteasome inhibitor-induced inclusions in PC12 cells by proteomic analysis. Acta Biochim Biophys Sin (Shanghai) 40:406–418. https://doi.org/10.1111/j.1745-7270.2008.00416.x
Li X, Zhang Y, Xie P, Piao J, Hu Y, Chang M, Liu T, Hu L (2010) Proteomic characterization of an isolated fraction of synthetic proteasome inhibitor (PSI)-induced inclusions in PC12 cells might offer clues to aggresomes as a cellular defensive response against proteasome inhibition by PSI. BMC Neurosci 11:95. https://doi.org/10.1186/1471-2202-11-95
Li Y, Jin P (2012) RNA-mediated neurodegeneration in fragile X-associated tremor/ataxia syndrome. Brain Res 1462:112–117. https://doi.org/10.1016/j.brainres.2012.02.057
Liebelt F, Vertegaal AC (2016) Ubiquitin-dependent and independent roles of SUMO in proteostasis. Am J Physiol Cell Physiol 311:C284–C296. https://doi.org/10.1152/ajpcell.00091.2016
Lilienbaum A (2013) Relationship between the proteasomal system and autophagy. International journal of biochemistry and molecular biology 4:1–26
Liu J, Koscielska KA, Cao Z, Hulsizer S, Grace N, Mitchell G, Nacey C, Githinji J, McGee J, Garcia-Arocena D, Hagerman RJ, Nolta J, Pessah IN, Hagerman PJ (2012) Signaling defects in iPSC-derived fragile X premutation neurons. Hum Mol Genet 21:3795–3805. https://doi.org/10.1093/hmg/dds207
Loesch DZ, Sherwell S, Kinsella G, Tassone F, Taylor A, Amor D, Sung S, Evans A (2012) Fragile X-associated tremor/ataxia phenotype in a male carrier of unmethylated full mutation in the FMR1 gene. Clin Genet 82:88–92. https://doi.org/10.1111/j.1399-0004.2011.01675.x
Lovtrup-Rein H, McEwen BS (1966) Isolation and fractionation of rat brain nuclei. J Cell Biol 30:405–415. https://doi.org/10.1083/jcb.30.2.405
Low P (2011) The role of ubiquitin-proteasome system in ageing. Gen Comp Endocrinol 172:39–43. https://doi.org/10.1016/j.ygcen.2011.02.005
Luo HB, Xia YY, Shu XJ, Liu ZC, Feng Y, Liu XH, Yu G, Yin G, Xiong YS, Zeng K, Jiang J, Ye K, Wang XC, Wang JZ (2014) SUMOylation at K340 inhibits tau degradation through deregulating its phosphorylation and ubiquitination. Proc Natl Acad Sci U S A 111:16586–16591. https://doi.org/10.1073/pnas.1417548111
Luo J, Gurung S, Lee L, Henley JM, Wilkinson KA, Guo C (2017) Increased SUMO-2/3-ylation mediated by SENP3 degradation is protective against cadmium-induced caspase 3-dependent cytotoxicity. J Toxicol Sci 42:529–538. https://doi.org/10.2131/jts.42.529
Mackenzie IR, Bigio EH, Ince PG, Geser F, Neumann M, Cairns NJ, Kwong LK, Forman MS, Ravits J, Stewart H, Eisen A, McClusky L, Kretzschmar HA, Monoranu CM, Highley JR, Kirby J, Siddique T, Shaw PJ, Lee VM, Trojanowski JQ (2007) Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol 61:427–434. https://doi.org/10.1002/ana.21147
Mackenzie IR, Frick P, Grasser FA, Gendron TF, Petrucelli L, Cashman NR, Edbauer D, Kremmer E, Prudlo J, Troost D, Neumann M (2015) Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol 130:845–861. https://doi.org/10.1007/s00401-015-1476-2
Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA (2001) Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet 10:2165–2170. https://doi.org/10.1093/hmg/10.19.2165
Marinello M, Werner A, Giannone M, Tahiri K, Alves S, Tesson C, den Dunnen W, Seeler JS, Brice A, Sittler A (2019) SUMOylation by SUMO2 is implicated in the degradation of misfolded ataxin-7 via RNF4 in SCA7 models. Dis Model Mech 12. https://doi.org/10.1242/dmm.036145
May S, Hornburg D, Schludi MH, Arzberger T, Rentzsch K, Schwenk BM, Grasser FA, Mori K, Kremmer E, Banzhaf-Strathmann J, Mann M, Meissner F, Edbauer D (2014) C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol 128:485–503. https://doi.org/10.1007/s00401-014-1329-4
Maziuk BF, Apicco DJ, Cruz AL, Jiang L, Ash PEA, da Rocha EL, Zhang C, Yu WH, Leszyk J, Abisambra JF, Li H, Wolozin B (2018) RNA binding proteins co-localize with small tau inclusions in tauopathy. Acta Neuropathol Commun 6:71. https://doi.org/10.1186/s40478-018-0574-5
McCormack A, Keating DJ, Chegeni N, Colella A, Wang JJ, Chataway T (2019) Abundance of Synaptic Vesicle-Related Proteins in Alpha-Synuclein-Containing Protein Inclusions Suggests a Targeted Formation Mechanism. Neurotox Res 35:883–897. https://doi.org/10.1007/s12640-019-00014-0
McEwen BS, Zigmond RE (1972) Isolation of Brain Cell Nuclei. In: Marks N, Rodnight R (eds) Research Methods in Neurochemistry, Springer, vol 1. Boston, MA, pp 139–161
Menzies FM, Fleming A, Caricasole A, Bento CF, Andrews SP, Ashkenazi A, Fullgrabe J, Jackson A, Jimenez Sanchez M, Karabiyik C, Licitra F, Lopez Ramirez A, Pavel M, Puri C, Renna M, Ricketts T, Schlotawa L, Vicinanza M, Won H, Zhu Y, Skidmore J, Rubinsztein DC (2017) Autophagy and Neurodegeneration: Pathogenic Mechanisms and Therapeutic Opportunities. Neuron 93:1015–1034. https://doi.org/10.1016/j.neuron.2017.01.022
Miteva M, Keusekotten K, Hofmann K, Praefcke GJ, Dohmen RJ (2010) Sumoylation as a signal for polyubiquitylation and proteasomal degradation. Sub-cellular biochemistry 54:195–214. https://doi.org/10.1007/978-1-4419-6676-6_16
Mitsui K, Nakayama H, Akagi T, Nekooki M, Ohtawa K, Takio K, Hashikawa T, Nukina N (2002) Purification of polyglutamine aggregates and identification of elongation factor-1alpha and heat shock protein 84 as aggregate-interacting proteins. J Neurosci 22:9267–9277
Mori F, Tanji K, Miki Y, Toyoshima Y, Sasaki H, Yoshida M, Kakita A, Takahashi H, Wakabayashi K (2018) Immunohistochemical localization of exoribonucleases (DIS3L2 and XRN1) in intranuclear inclusion body disease. Neurosci Lett 662:389–394. https://doi.org/10.1016/j.neulet.2017.10.061
Myeku N, Clelland CL, Emrani S, Kukushkin NV, Yu WH, Goldberg AL, Duff KE (2016) Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat Med 22:46–53. https://doi.org/10.1038/nm.4011
Napoli E, Ross-Inta C, Wong S, Omanska-Klusek A, Barrow C, Iwahashi C, Garcia-Arocena D, Sakaguchi D, Berry-Kravis E, Hagerman R, Hagerman PJ, Giulivi C (2011) Altered zinc transport disrupts mitochondrial protein processing/import in fragile X-associated tremor/ataxia syndrome. Hum Mol Genet 20:3079–3092. https://doi.org/10.1093/hmg/ddr211
Nemes Z, Petrovski G, Aerts M, Sergeant K, Devreese B, Fesus L (2009) Transglutaminase-mediated intramolecular cross-linking of membrane-bound alpha-synuclein promotes amyloid formation in Lewy bodies. J Biol Chem 284:27252–27264. https://doi.org/10.1074/jbc.M109.033969
Nielsen SV, Poulsen EG, Rebula CA, Hartmann-Petersen R (2014) Protein quality control in the nucleus. Biomolecules 4:646–661. https://doi.org/10.3390/biom4030646
Norman JM, Cohen GM, Bampton ET (2010) The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy 6:1042–1056. https://doi.org/10.4161/auto.6.8.13337
Oh SY, He F, Krans A, Frazer M, Taylor JP, Paulson HL, Todd PK (2015) RAN translation at CGG repeats induces ubiquitin proteasome system impairment in models of fragile X-associated tremor ataxia syndrome. Hum Mol Genet 24:4317–4326. https://doi.org/10.1093/hmg/ddv165
O'Rourke JG, Gareau JR, Ochaba J, Song W, Rasko T, Reverter D, Lee J, Monteys AM, Pallos J, Mee L, Vashishtha M, Apostol BL, Nicholson TP, Illes K, Zhu YZ, Dasso M, Bates GP, Difiglia M, Davidson B, Wanker EE, Marsh JL, Lima CD, Steffan JS, Thompson LM (2013) SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Rep 4:362–375. https://doi.org/10.1016/j.celrep.2013.06.034
Ottis P, Koppe K, Onisko B, Dynin I, Arzberger T, Kretzschmar H, Requena JR, Silva CJ, Huston JP, Korth C (2012) Human and rat brain lipofuscin proteome. Proteomics 12:2445–2454. https://doi.org/10.1002/pmic.201100668
Pankiv S, Lamark T, Bruun JA, Overvatn A, Bjorkoy G, Johansen T (2010) Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol Chem 285:5941–5953. https://doi.org/10.1074/jbc.M109.039925
Pedraza L, Fidler L, Staugaitis SM, Colman DR (1997) The active transport of myelin basic protein into the nucleus suggests a regulatory role in myelination. Neuron 18:579–589
Petersen TW, Brent Harrison C, Horner DN, van den Engh G (2012) Flow cytometric characterization of marine microbes. Methods 57:350–358. https://doi.org/10.1016/j.ymeth.2012.07.001
Pountney DL, Huang Y, Burns RJ, Haan E, Thompson PD, Blumbergs PC, Gai WP (2003) SUMO-1 marks the nuclear inclusions in familial neuronal intranuclear inclusion disease. Exp Neurol 184:436–446. https://doi.org/10.1016/j.expneurol.2003.07.004
Pountney DL, Raftery MJ, Chegini F, Blumbergs PC, Gai WP (2008) NSF, Unc-18-1, dynamin-1 and HSP90 are inclusion body components in neuronal intranuclear inclusion disease identified by anti-SUMO-1-immunocapture. Acta Neuropathol 116:603–614. https://doi.org/10.1007/s00401-008-0437-4
Pountney DL, Treweek TM, Chataway T, Huang Y, Chegini F, Blumbergs PC, Raftery MJ, Gai WP (2005) Alpha B-crystallin is a major component of glial cytoplasmic inclusions in multiple system atrophy. Neurotox Res 7:77–85
Pretto D, Yrigollen CM, Tang HT, Williamson J, Espinal G, Iwahashi CK, Durbin-Johnson B, Hagerman RJ, Hagerman PJ, Tassone F (2014) Clinical and molecular implications of mosaicism in FMR1 full mutations. Front Genet 5:318. https://doi.org/10.3389/fgene.2014.00318
Pretto DI, Hunsaker MR, Cunningham CL, Greco CM, Hagerman RJ, Noctor SC, Hall DA, Hagerman PJ, Tassone F (2013) Intranuclear inclusions in a fragile X mosaic male. Transl Neurodegener 2:10. https://doi.org/10.1186/2047-9158-2-10
Qurashi A, Li W, Zhou JY, Peng J, Jin P (2011) Nuclear accumulation of stress response mRNAs contributes to the neurodegeneration caused by Fragile X premutation rCGG repeats. PLoS Genet 7:e1002102. https://doi.org/10.1371/journal.pgen.1002102
Rhoades E (ed) (2018) Intrinsically Disordered Proteins, vol 611. Academic Press, Cambridge, MA
Robin G, Lopez JR, Espinal GM, Hulsizer S, Hagerman PJ, Pessah IN (2017) Calcium dysregulation and Cdk5-ATM pathway involved in a mouse model of fragile X-associated tremor/ataxia syndrome. Hum Mol Genet 26:2649–2666. https://doi.org/10.1093/hmg/ddx148
Rodriguez CM, Todd PK (2019) New pathologic mechanisms in nucleotide repeat expansion disorders. Neurobiol Dis 130:104515. https://doi.org/10.1016/j.nbd.2019.104515
Roshchina VV (2012) Vital Autofluorescence: Application to the Study of Plant Living Cells. International Journal of Spectroscopy 2012:14. https://doi.org/10.1155/2012/124672
Ross-Inta C, Omanska-Klusek A, Wong S, Barrow C, Garcia-Arocena D, Iwahashi C, Berry-Kravis E, Hagerman RJ, Hagerman PJ, Giulivi C (2010) Evidence of mitochondrial dysfunction in fragile X-associated tremor/ataxia syndrome. Biochem J 429:545–552. https://doi.org/10.1042/BJ20091960
Rotondo R, Moschini R, Renzone G, Tuccinardi T, Balestri F, Cappiello M, Scaloni A, Mura U, Del-Corso A (2016) Human carbonyl reductase 1 as efficient catalyst for the reduction of glutathionylated aldehydes derived from lipid peroxidation. Free Radic Biol Med 99:323–332. https://doi.org/10.1016/j.freeradbiomed.2016.08.015
Sagne C, Isambert MF, Henry JP, Gasnier B (1996) SDS-resistant aggregation of membrane proteins: application to the purification of the vesicular monoamine transporter. Biochem J 316(Pt 3):825–831. https://doi.org/10.1042/bj3160825
Sato K, Murakami T, Hamakawa Y, Kamada H, Nagano I, Shoji M, Takata H, Nobukuni K, Ihara Y, Namba R, Hayabara T, Hirose S, Abe K (2002) Selective colocalization of transglutaminase-like activity in ubiquitinated intranuclear inclusions of hereditary dentatorubral-pallidoluysian atrophy. Brain Res 952:327–330. https://doi.org/10.1016/s0006-8993(02)03244-4
Schludi MH, Becker L, Garrett L, Gendron TF, Zhou Q, Schreiber F, Popper B, Dimou L, Strom TM, Winkelmann J, von Thaden A, Rentzsch K, May S, Michaelsen M, Schwenk BM, Tan J, Schoser B, Dieterich M, Petrucelli L, Holter SM, Wurst W, Fuchs H, Gailus-Durner V, de Angelis MH, Klopstock T, Arzberger T, Edbauer D (2017) Spinal poly-GA inclusions in a C9orf72 mouse model trigger motor deficits and inflammation without neuron loss. Acta Neuropathol 134:241–254. https://doi.org/10.1007/s00401-017-1711-0
Sellier C, Buijsen RAM, He F, Natla S, Jung L, Tropel P, Gaucherot A, Jacobs H, Meziane H, Vincent A, Champy MF, Sorg T, Pavlovic G, Wattenhofer-Donze M, Birling MC, Oulad-Abdelghani M, Eberling P, Ruffenach F, Joint M, Anheim M, Martinez-Cerdeno V, Tassone F, Willemsen R, Hukema RK, Viville S, Martinat C, Todd PK, Charlet-Berguerand N (2017) Translation of Expanded CGG Repeats into FMRpolyG Is Pathogenic and May Contribute to Fragile X Tremor Ataxia Syndrome. Neuron 93:331–347. https://doi.org/10.1016/j.neuron.2016.12.016
Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F, Alunni V, Moine H, Thibault C, Page A, Tassone F, Willemsen R, Disney MD, Hagerman PJ, Todd PK, Charlet-Berguerand N (2013) Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep 3:869–880. https://doi.org/10.1016/j.celrep.2013.02.004
Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, Schneider A, Richard S, Willemsen R, Elliott DJ, Hagerman PJ, Charlet-Berguerand N (2010) Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J 29:1248–1261. https://doi.org/10.1038/emboj.2010.21
Seyfried NT, Gozal YM, Donovan LE, Herskowitz JH, Dammer EB, Xia Q, Ku L, Chang J, Duong DM, Rees HD, Cooper DS, Glass JD, Gearing M, Tansey MG, Lah JJ, Feng Y, Levey AI, Peng J (2012) Quantitative analysis of the detergent-insoluble brain proteome in frontotemporal lobar degeneration using SILAC internal standards. J Proteome Res 11:2721–2738. https://doi.org/10.1021/pr2010814
Sinigalliano CD, Winshell J, Guerrero MA, Scorzetti G, Fell JW, Eaton RW, Brand L, Rein KS (2009) Viable cell sorting of dinoflagellates by multiparametric flow cytometry. Phycologia 48:249–257
Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J (2007) RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron 55:565–571. https://doi.org/10.1016/j.neuron.2007.07.021
Song G, Napoli E, Wong S, Hagerman R, Liu S, Tassone F, Giulivi C (2016) Altered redox mitochondrial biology in the neurodegenerative disorder fragile X-tremor/ataxia syndrome: use of antioxidants in precision medicine. Mol Med 22:548–559. https://doi.org/10.2119/molmed.2016.00122
Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, Pandolfi PP, Thompson LM, Marsh JL (2004) SUMO modification of Huntingtin and Huntington's disease pathology. Science 304:100–104. https://doi.org/10.1126/science.1092194
Tanaka G, Yamanaka T, Furukawa Y, Kajimura N, Mitsuoka K, Nukina N (2019) Biochemical and morphological classification of disease-associated alpha-synuclein mutants aggregates. Biochem Biophys Res Commun 508:729–734. https://doi.org/10.1016/j.bbrc.2018.11.200
Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ (2000) Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet 66:6–15. https://doi.org/10.1086/302720
Tassone F, Iwahashi C, Hagerman PJ (2004) FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS). RNA Biol 1:103–105. https://doi.org/10.4161/rna.1.2.1035
Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS (1996) Identification of a (CUG) n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res 24:4407–4414. https://doi.org/10.1093/nar/24.22.4407
Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, Renoux AJ, Chen KC, Scaglione KM, Basrur V, Elenitoba-Johnson K, Vonsattel JP, Louis ED, Sutton MA, Taylor JP, Mills RE, Charlet-Berguerand N, Paulson HL (2013) CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78:440–455. https://doi.org/10.1016/j.neuron.2013.03.026
Tsai AM, van Zanten JH, Betenbaugh MJ (1998) I. Study of protein aggregation due to heat denaturation: A structural approach using circular dichroism spectroscopy, nuclear magnetic resonance, and static light scattering. Biotechnology and bioengineering 59:273–280
Tsutakawa SE, Yan C, Xu X, Weinacht CP, Freudenthal BD, Yang K, Zhuang Z, Washington MT, Tainer JA, Ivanov I (2015) Structurally distinct ubiquitin- and sumo-modified PCNA: implications for their distinct roles in the DNA damage response. Structure 23:724–733. https://doi.org/10.1016/j.str.2015.02.008
van Wijk SJ, Muller S, Dikic I (2011) Shared and unique properties of ubiquitin and SUMO interaction networks in DNA repair. Genes Dev 25:1763–1769. https://doi.org/10.1101/gad.17593511
Walker C, Herranz-Martin S, Karyka E, Liao C, Lewis K, Elsayed W, Lukashchuk V, Chiang SC, Ray S, Mulcahy PJ, Jurga M, Tsagakis I, Iannitti T, Chandran J, Coldicott I, De Vos KJ, Hassan MK, Higginbottom A, Shaw PJ, Hautbergue GM, Azzouz M, El-Khamisy SF (2017) C9orf72 expansion disrupts ATM-mediated chromosomal break repair. Nat Neurosci 20:1225–1235. https://doi.org/10.1038/nn.4604
Willemsen R, Hoogeveen-Westerveld M, Reis S, Holstege J, Severijnen LA, Nieuwenhuizen IM, Schrier M, van Unen L, Tassone F, Hoogeveen AT, Hagerman PJ, Mientjes EJ, Oostra BA (2003) The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum Mol Genet 12:949–959. https://doi.org/10.1093/hmg/ddg114
Wooten MW, Hu X, Babu JR, Seibenhener ML, Geetha T, Paine MG, Wooten MC (2006) Signaling, polyubiquitination, trafficking, and inclusions: sequestosome 1/p62's role in neurodegenerative disease. J Biomed Biotechnol 2006:62079. https://doi.org/10.1155/JBB/2006/62079
Xia Q, Liao L, Cheng D, Duong DM, Gearing M, Lah JJ, Levey AI, Peng J (2008) Proteomic identification of novel proteins associated with Lewy bodies. Front Biosci 13:3850–3856
Xiao Y, Pollack D, Nieves E, Winchell A, Callaway M, Vigodner M (2015) Can your protein be sumoylated? A quick summary and important tips to study SUMO-modified proteins. Anal Biochem 477:95–97. https://doi.org/10.1016/j.ab.2014.11.006
Yang G, Liu C, Chen SH, Kassab MA, Hoff JD, Walter NG, Yu X (2018) Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res 46:3446–3457. https://doi.org/10.1093/nar/gky088
Yang L, Zhou Y, Zhu S, Huang T, Wu L, Yan X (2012) Detection and quantification of bacterial autofluorescence at the single-cell level by a laboratory-built high-sensitivity flow cytometer. Anal Chem 84:1526–1532. https://doi.org/10.1021/ac2031332
Yang WY, Larios E, Gruebele M (2003) On the extended beta-conformation propensity of polypeptides at high temperature. J Am Chem Soc 125:16220–16227. https://doi.org/10.1021/ja0360081
Yang Y, He Y, Wang X, Liang Z, He G, Zhang P, Zhu H, Xu N, Liang S (2017) Protein SUMOylation modification and its associations with disease. Open Biol 7. https://doi.org/10.1098/rsob.170167
Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU (2006) Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 8:1124–1132. https://doi.org/10.1038/ncb1482
Yun M, Choi AJ, Lee YC, Kong M, Sung JY, Kim SS, Eun YG (2018) Carbonyl reductase 1 is a new target to improve the effect of radiotherapy on head and neck squamous cell carcinoma. J Exp Clin Cancer Res 37:264. https://doi.org/10.1186/s13046-018-0942-9
Yun SM, Cho SJ, Song JC, Song SY, Jo SA, Jo C, Yoon K, Tanzi RE, Choi EJ, Koh YH (2013) SUMO1 modulates Abeta generation via BACE1 accumulation. Neurobiol Aging 34:650–662. https://doi.org/10.1016/j.neurobiolaging.2012.08.005
Zanon A, Rakovic A, Blankenburg H, Doncheva NT, Schwienbacher C, Serafin A, Alexa A, Weichenberger CX, Albrecht M, Klein C, Hicks AA, Pramstaller PP, Domingues FS, Pichler I (2013) Profiling of Parkin-binding partners using tandem affinity purification. PLoS One 8:e78648. https://doi.org/10.1371/journal.pone.0078648
Zatloukal K, Stumptner C, Fuchsbichler A, Heid H, Schnoelzer M, Kenner L, Kleinert R, Prinz M, Aguzzi A, Denk H (2002) p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. The American journal of pathology 160:255–263. https://doi.org/10.1016/s0002-9440(10)64369-6
Zhang XD, Goeres J, Zhang H, Yen TJ, Porter AC, Matunis MJ (2008) SUMO-2/3 modification and binding regulate the association of CENP-E with kinetochores and progression through mitosis. Mol Cell 29:729–741. https://doi.org/10.1016/j.molcel.2008.01.013
Zheng H, Yuan C, Zhang H, Chen Y, Zhang H (2019) The tissue- and developmental stage-specific involvement of autophagy genes in aggrephagy. Autophagy. 2019:1–11. https://doi.org/10.1080/15548627.2019.1632121. Epub ahead of print.
Zhou Y, Gu G, Goodlett DR, Zhang T, Pan C, Montine TJ, Montine KS, Aebersold RH, Zhang J (2004) Analysis of alpha-synuclein-associated proteins by quantitative proteomics. J Biol Chem 279:39155–39164. https://doi.org/10.1074/jbc.M405456200
Acknowledgements
The authors wish to dedicate this paper to the memory of Christine Iwahashi, whose early investigations of the isolated inclusions provided a foundation for the current work.
Tissue specimens were obtained from the National Institutes of Health (NIH) Brain & Tissue Repository-California, Human Brain & Spinal Fluid Resource Center, VA West Los Angeles Medical Center, Los Angeles, California, which is supported in part by NIH and the US Department of Veterans Affairs. Tissue specimens were also obtained from the Alzheimer’s Disease Center Biorepository at University of California, Davis (UCD), which is supported in part by the Neuropathology Core Grant P30 AG010129. Mouse tissue specimens were collected with assistance from Shane Antrobus at the UCD School of Veterinary Medicine. Assistance was provided by Ingrid Brust-Mascher at the Health Sciences District Advanced Imaging Facility at UCD using the Leica SP8 STED 3x. LC-MS/MS sample preparation and data acquisition was performed at the UCD Proteomics Facility. This project was supported by the UCD Flow Cytometry Shared Resource Laboratory with funding from the NCI P30 CA093373 (Cancer Center) and S10 OD018223 (Astrios Cell Sorter) grants, with technical assistance from Mr. Jonathan Van Dyke.
Funding
This study was partially funded by the Yearlong Exposure to Advanced Research training program (LM) under grant number T32OD010931 and the Marjorie and Charles Elliott Fellowship offered by the UCD Integrative Genetics and Genomics Graduate Group. This work was also supported by NIH grants R01 HD036071 (RJH), R01 GM113929 (PJH), P01 ES011269 (INP), R01 ES030318 (INP), R01 NS107131 (VM-C), and by NSF grant 1840842 (INP).
Author information
Authors and Affiliations
Contributions
LM, INP, AWH, and PJH were responsible for intellectual and experimental concept and design. Mass spectrometry experimental design and data collection were performed by AWH. Two western blots were performed by GE. FMRpolyG positive control construct design and creation was performed by JR. Fluorescence activated cell sorting was performed by BM. Frontal cortex dissection for some human samples was performed by VM-C. Mice were managed and provided by INP. FXTAS patient diagnosis was confirmed by RJH. All other material preparation, data collection, and analysis were performed by LM with support from PJH. The first draft of the manuscript was written by LM and all authors contributed to subsequent versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Human Participants: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee (UCD Institutional Review Board; protocol #215292) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. Informed consent was obtained from all individual participants or their family designates included in the study.
Animals: All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted (UCD Institutional Animal Care and Use Committee; protocol #20431).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
This file contains additional methods, supplementary figures, figure legends, and tables in support of the main text. (DOCX 5244 kb)
Additional file 2:
This file contains raw data for the two MS analyses in the tabs labeled “Protein groups”; analyzed data to calculate relative percent abundances of identified proteins in the tabs labeled “Relative abundance”; and a list of the 176 proteins found enriched in FXTAS inclusions and used for Fig. 3 in the tab labeled “176 enriched inclusion proteins. (XLSX 2720 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ma, L., Herren, A.W., Espinal, G. et al. Composition of the Intranuclear Inclusions of Fragile X-associated Tremor/Ataxia Syndrome. acta neuropathol commun 7, 143 (2019). https://doi.org/10.1186/s40478-019-0796-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40478-019-0796-1