Abstract
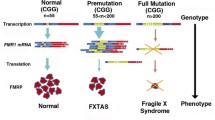
Fragile X-associated tremor/ataxia syndrome (FXTAS) is an inherited neurodegenerative disorder manifesting in carriers of 55 to 200 CGG repeats in the 5′ untranslated region (UTR) of the fragile X mental retardation gene (FMR1). FXTAS is characterized by enhanced FMR1 transcription and the accumulation of CGG repeat-containing FMR1 messenger RNA in nuclear foci, while the FMRP protein expression levels remain normal or moderately low. The neuropathological hallmark in FXTAS is the presence of intranuclear, ubiquitin-positive inclusions that also contain FMR1 transcript. Yet, the complete protein complement of FXTAS inclusions and the molecular events that trigger neuronal death in FXTAS remain unclear. In this review, we present the two most accepted toxicity mechanisms described so far, namely RNA gain-of-function and protein gain-of-function by means of repeat-associated non-AUG translation, and discuss current experimental and computational strategies to better understand FXTAS pathogenesis. Finally, we review the current perspectives for drug development with disease-modifying potential for FXTAS.

Similar content being viewed by others
References
Hagerman PJ, Hagerman RJ. Fragile X-associated tremor/ataxia syndrome. Ann N Y Acad Sci. 2015;1338:58–70.
Brown SSG, Stanfield AC. Fragile X premutation carriers: a systematic review of neuroimaging findings. J Neurol Sci. 2015;352:19–28.
Rodriguez-Revenga L, Madrigal I, Pagonabarraga J, Xunclà M, Badenas C, Kulisevsky J, et al. Penetrance of FMR1 premutation associated pathologies in fragile X syndrome families. Eur J Hum Genet EJHG. 2009;17:1359–62.
Cronister A, Teicher J, Rohlfs EM, Donnenfeld A, Hallam S. Prevalence and instability of fragile X alleles: implications for offering fragile X prenatal diagnosis. Obstet Gynecol. 2008;111:596–601.
Usdin K, Hayward BE, Kumari D, Lokanga RA, Sciascia N, Zhao X-N. Repeat-mediated genetic and epigenetic changes at the FMR1 locus in the Fragile X-related disorders. Front Genet. 2014;5:226.
Leehey MA. Fragile X-associated tremor/ataxia syndrome: clinical phenotype, diagnosis, and treatment. J Investig Med Off Publ Am Fed Clin Res. 2009;57:830–6.
Hagerman RJ, Hall DA, Coffey S, Leehey M, Bourgeois J, Gould J, et al. Treatment of fragile X-associated tremor ataxia syndrome (FXTAS) and related neurological problems. Clin Interv Aging. 2008;3:251–62.
Polussa J, Schneider A, Hagerman R. Molecular advances leading to treatment implications for fragile X premutation carriers. Brain Disord Ther. 2014;3.
Kenneson A, Zhang F, Hagedorn CH, Warren ST. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum Mol Genet. 2001;10:1449–54.
Primerano B, Tassone F, Hagerman RJ, Hagerman P, Amaldi F, Bagni C. Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. RNA N Y N. 2002;8:1482–8.
Tassone F, Hagerman RJ, Taylor AK, Mills JB, Harris SW, Gane LW, et al. Clinical involvement and protein expression in individuals with the FMR1 premutation. Am J Med Genet. 2000;91:144–52.
Brouwer JR, Huizer K, Severijnen L-A, Hukema RK, Berman RF, Oostra BA, et al. CGG-repeat length and neuropathological and molecular correlates in a mouse model for fragile X-associated tremor/ataxia syndrome. J Neurochem. 2008;107:1671–82.
Ludwig AL, Espinal GM, Pretto DI, Jamal AL, Arque G, Tassone F, et al. CNS expression of murine fragile X protein (FMRP) as a function of CGG-repeat size. Hum Mol Genet. 2014;23:3228–38.
Lozano R, Rosero CA, Hagerman RJ. Fragile X spectrum disorders. Intractable Rare Dis Res. 2014;3:134–46.
Gokden M, Al-Hinti JT, Harik SI. Peripheral nervous system pathology in fragile X tremor/ataxia syndrome (FXTAS). Neuropathol Off J Jpn Soc Neuropathol. 2009;29:280–4.
Greco CM, Berman RF, Martin RM, Tassone F, Schwartz PH, Chang A, et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS). Brain J Neurol. 2006;129:243–55.
Greco CM, Soontrapornchai K, Wirojanan J, Gould JE, Hagerman PJ, Hagerman RJ. Testicular and pituitary inclusion formation in fragile X associated tremor/ataxia syndrome. J Urol. 2007;177:1434–7.
Hunsaker MR, Greco CM, Spath MA, Smits APT, Navarro CS, Tassone F, et al. Widespread non-central nervous system organ pathology in fragile X premutation carriers with fragile X-associated tremor/ataxia syndrome and CGG knock-in mice. Acta Neuropathol (Berl). 2011;122:467–79.
Ariza J, Rogers H, Monterrubio A, Reyes-Miranda A, Hagerman PJ, Martínez-Cerdeño V. A majority of FXTAS cases present with intranuclear inclusions within purkinje cells. Cerebellum Lond Engl. 2016.
Iwahashi CK, Yasui DH, An H-J, Greco CM, Tassone F, Nannen K, et al. Protein composition of the intranuclear inclusions of FXTAS. Brain J Neurol. 2006;129:256–71.
Greco CM, Hagerman RJ, Tassone F, Chudley AE, Del Bigio MR, Jacquemont S, et al. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain J Neurol. 2002;125:1760–71.
Wojciechowska M, Krzyzosiak WJ. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum Mol Genet. 2011;20:3811–21.
Ranum LPW, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–77.
Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–61.
Bekenstein U, Soreq H. Heterogeneous nuclear ribonucleoprotein A1 in health and neurodegenerative disease: from structural insights to post-transcriptional regulatory roles. Mol Cell Neurosci. 2013;56:436–46.
Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, et al. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–71.
Muslimov IA, Patel MV, Rose A, Tiedge H. Spatial code recognition in neuronal RNA targeting: role of RNA-hnRNP A2 interactions. J Cell Biol. 2011;194:441–57.
Iijima T, Wu K, Witte H, Hanno-Iijima Y, Glatter T, Richard S, et al. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011;147:1601–14.
Klein ME, Younts TJ, Castillo PE, Jordan BA. RNA-binding protein Sam68 controls synapse number and local β-actin mRNA metabolism in dendrites. Proc Natl Acad Sci U S A. 2013;110:3125–30.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33.
Sellier C, Freyermuth F, Tabet R, Tran T, He F, Ruffenach F, et al. Sequestration of DROSHA and DGCR8 by expanded CGG RNA repeats alters microRNA processing in fragile X-associated tremor/ataxia syndrome. Cell Rep. 2013;3:869–80.
Alvarez-Mora MI, Rodriguez-Revenga L, Madrigal I, Torres-Silva F, Mateu-Huertas E, Lizano E, et al. MicroRNA expression profiling in blood from fragile X-associated tremor/ataxia syndrome patients. Genes Brain Behav. 2013;12:595–603.
Tan H, Poidevin M, Li H, Chen D, Jin P. MicroRNA-277 modulates the neurodegeneration caused by Fragile X premutation rCGG repeats. PLoS Genet. 2012;8, e1002681.
Fénelon K, Mukai J, Xu B, Hsu P-K, Drew LJ, Karayiorgou M, et al. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc Natl Acad Sci U S A. 2011;108:4447–52.
Luhur A, Chawla G, Wu Y-C, Li J, Sokol NS. Drosha-independent DGCR8/Pasha pathway regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2014;111:1421–6.
Grigsby J, Cornish K, Hocking D, Kraan C, Olichney JM, Rivera SM, et al. The cognitive neuropsychological phenotype of carriers of the FMR1 premutation. J Neurodev Disord. 2014;6:28.
Seritan AL, Ortigas M, Seritan S, Bourgeois JA, Hagerman RJ. Psychiatric disorders associated with FXTAS. Curr Psychiatry Rev. 2013;9:59–64.
Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, et al. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–64.
Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, et al. Pur α binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–64.
Hunt D, Leventer RJ, Simons C, Taft R, Swoboda KJ, Gawne-Cain M, et al. Whole exome sequencing in family trios reveals de novo mutations in PURA as a cause of severe neurodevelopmental delay and learning disability. J Med Genet. 2014;51:806–13.
Lalani SR, Zhang J, Schaaf CP, Brown CW, Magoulas P, Tsai AC-H, et al. Mutations in PURA cause profound neonatal hypotonia, seizures, and encephalopathy in 5q31.3 microdeletion syndrome. Am J Hum Genet. 2014;95:579–83.
He F, Krans A, Freibaum BD, Taylor JP, Todd PK. TDP-43 suppresses CGG repeat-induced neurotoxicity through interactions with HnRNP A2/B1. Hum Mol Genet. 2014;23:5036–51.
O’Dwyer JP, Clabby C, Crown J, Barton DE, Hutchinson M. Fragile X-associated tremor/ataxia syndrome presenting in a woman after chemotherapy. Neurology. 2005;65:331–2.
Paul R, Pessah IN, Gane L, Ono M, Hagerman PJ, Brunberg JA, et al. Early onset of neurological symptoms in fragile X premutation carriers exposed to neurotoxins. Neurotoxicology. 2010;31:399–402.
Siprashvili Z, Webster DE, Kretz M, Johnston D, Rinn JL, Chang HY, et al. Identification of proteins binding coding and non-coding human RNAs using protein microarrays. BMC Genomics. 2012;13:633.
Simon MD. Capture hybridization analysis of RNA targets (CHART). Curr Protoc Mol Biol Ed. Frederick M Ausubel Al Chapter 21, Unit 21.25. 2013.
Tsai BP, Wang X, Huang L, Waterman ML. Quantitative profiling of in vivo-assembled RNA-protein complexes using a novel integrated proteomic approach. Mol Cell Proteomics MCP. 2011;10:M110.007385.
Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP). J Vis Exp JoVE. 2012.
Urbanek MO, Krzyzosiak WJ. RNA FISH for detecting expanded repeats in human diseases. Methods San Diego Calif. 2015.
Agostini F, Zanzoni A, Klus P, Marchese D, Cirillo D, Tartaglia GG. catRAPID omics: a web server for large-scale prediction of protein-RNA interactions. Bioinforma Oxf Engl. 2013a.
Bellucci M, Agostini F, Masin M, Tartaglia GG. Predicting protein associations with long noncoding RNAs. Nat Methods. 2011;8:444–5.
Stawiski EW, Gregoret LM, Mandel-Gutfreund Y. Annotating nucleic acid-binding function based on protein structure. J Mol Biol. 2003;326:1065–79.
Wu T, Wang J, Liu C, Zhang Y, Shi B, Zhu X, et al. NPInter: the noncoding RNAs and protein related biomacromolecules interaction database. Nucleic Acids Res. 2006;34:D150–2.
Agostini F, Cirillo D, Bolognesi B, Tartaglia GG. X-inactivation: quantitative predictions of protein interactions in the Xist network. Nucleic Acids Res. 2013;41, e31.
Cirillo D, Agostini F, Klus P, Marchese D, Rodriguez S, Bolognesi B, et al. Neurodegenerative diseases: quantitative predictions of protein-RNA interactions. RNA N Y N. 2013;19:129–40.
Cirillo D, Marchese D, Agostini F, Livi CM, Botta-Orfila T, Tartaglia GG. Constitutive patterns of gene expression regulated by RNA-binding proteins. Genome Biol. 2014;15:R13.
Zanzoni A, Marchese D, Agostini F, Bolognesi B, Cirillo D, Botta-Orfila M, et al. Principles of self-organization in biological pathways: a hypothesis on the autogenous association of alpha-synuclein. Nucleic Acids Res. 2013. gkt794.
Garcia-Arocena D, Hagerman PJ. Advances in understanding the molecular basis of FXTAS. Hum Mol Genet. 2010;19:R83–9.
Loomis EW, Sanz LA, Chédin F, Hagerman PJ. Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet. 2014;10, e1004294.
Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108:260–5.
Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, et al. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron. 2013;78:440–55.
Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin W-L, Dejesus-Hernandez M, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–46.
Mori K, Weng S-M, Arzberger T, May S, Rentzsch K, Kremmer E, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–8.
Zu T, Liu Y, Bañez-Coronel M, Reid T, Pletnikova O, Lewis J, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci U S A. 2013;110:E4968–77.
Bañez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, et al. RAN translation in Huntington disease. Neuron. 2015;88:667–77.
Buijsen RAM, Sellier C, Severijnen L-AWFM, Oulad-Abdelghani M, Verhagen RFM, Berman RF, et al. FMRpolyG-positive inclusions in CNS and non-CNS organs of a fragile X premutation carrier with fragile X-associated tremor/ataxia syndrome. Acta Neuropathol Commun. 2014;2:162.
Buijsen RAM, Visser JA, Kramer P, Severijnen EAWFM, Gearing M, Charlet-Berguerand N, et al. Presence of inclusions positive for polyglycine containing protein, FMRpolyG, indicates that repeat-associated non-AUG translation plays a role in fragile X-associated primary ovarian insufficiency. Hum Reprod Oxf Engl. 2016;31:158–68.
Gendron TF, Bieniek KF, Zhang Y-J, Jansen-West K, Ash PEA, Caulfield T, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol (Berl). 2013;126:829–44.
Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, et al. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet. 2007;16:3174–87.
Oh SY, He F, Krans A, Frazer M, Taylor JP, Paulson HL, et al. RAN translation at CGG repeats induces ubiquitin proteasome system impairment in models of fragile X-associated tremor ataxia syndrome. Hum Mol Genet. 2015;24:4317–26.
Ohnishi S, Kamikubo H, Onitsuka M, Kataoka M, Shortle D. Conformational preference of polyglycine in solution to elongated structure. J Am Chem Soc. 2006;128:16338–44.
Oma Y, Kino Y, Sasagawa N, Ishiura S. Intracellular localization of homopolymeric amino acid-containing proteins expressed in mammalian cells. J Biol Chem. 2004;279:21217–22.
Oma Y, Kino Y, Sasagawa N, Ishiura S. Comparative analysis of the cytotoxicity of homopolymeric amino acids. Biochim Biophys Acta. 2005;1748:174–9.
Oma Y, Kino Y, Toriumi K, Sasagawa N, Ishiura S. Interactions between homopolymeric amino acids (HPAAs). Protein Sci Publ Protein Soc. 2007;16:2195–204.
Ciryam P, Tartaglia GG, Morimoto RI, Dobson CM, Vendruscolo M. Widespread aggregation and neurodegenerative diseases are associated with supersaturated proteins. Cell Rep. 2013;5:781–90.
Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78.
Mannini B, Mulvihill E, Sgromo C, Cascella R, Khodarahmi R, Ramazzotti M, et al. Toxicity of protein oligomers is rationalized by a function combining size and surface hydrophobicity. ACS Chem Biol. 2014;9:2309–17.
Campioni S, Mannini B, Zampagni M, Pensalfini A, Parrini C, Evangelisti E, et al. A causative link between the structure of aberrant protein oligomers and their toxicity. Nat Chem Biol. 2010;6:140–7.
De Baets G, Van Doorn L, Rousseau F, Schymkowitz J. Increased aggregation is more frequently associated to human disease-associated mutations than to neutral polymorphisms. PLoS Comput Biol. 2015;11, e1004374.
Lindersson E, Beedholm R, Højrup P, Moos T, Gai W, Hendil KB, et al. Proteasomal inhibition by alpha-synuclein filaments and oligomers. J Biol Chem. 2004;279:12924–34.
Díaz-Hernández M, Valera AG, Morán MA, Gómez-Ramos P, Alvarez-Castelao B, Castaño JG, et al. Inhibition of 26S proteasome activity by huntingtin filaments but not inclusion bodies isolated from mouse and human brain. J Neurochem. 2006;98:1585–96.
Ortega Z, Díaz-Hernández M, Maynard CJ, Hernández F, Dantuma NP, Lucas JJ. Acute polyglutamine expression in inducible mouse model unravels ubiquitin/proteasome system impairment and permanent recovery attributable to aggregate formation. J Neurosci Off J Soc Neurosci. 2010;30:3675–88.
Tseng BP, Green KN, Chan JL, Blurton-Jones M, LaFerla FM. Abeta inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiol Aging. 2008;29:1607–18.
Tang G, Perng MD, Wilk S, Quinlan R, Goldman JE. Oligomers of mutant glial fibrillary acidic protein (GFAP) Inhibit the proteasome system in alexander disease astrocytes, and the small heat shock protein alphaB-crystallin reverses the inhibition. J Biol Chem. 2010;285:10527–37.
Deriziotis P, André R, Smith DM, Goold R, Kinghorn KJ, Kristiansen M, et al. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. EMBO J. 2011;30:3065–77.
McKinnon C, Goold R, Andre R, Devoy A, Ortega Z, Moonga J, et al. Prion-mediated neurodegeneration is associated with early impairment of the ubiquitin-proteasome system. Acta Neuropathol (Berl.) 2015.
Woerner AC, Frottin F, Hornburg D, Feng LR, Meissner F, Patra M, et al. Cytoplasmic protein aggregates interfere with nucleocytoplasmic transport of protein and RNA. Science. 2016;351:173–6.
Kaplan ES, Cao Z, Hulsizer S, Tassone F, Berman RF, Hagerman PJ, et al. Early mitochondrial abnormalities in hippocampal neurons cultured from Fmr1 pre-mutation mouse model. J Neurochem. 2012;123:613–21.
Napoli E, Ross-Inta C, Wong S, Omanska-Klusek A, Barrow C, Iwahashi C, et al. Altered zinc transport disrupts mitochondrial protein processing/import in fragile X-associated tremor/ataxia syndrome. Hum Mol Genet. 2011;20:3079–92.
Ross-Inta C, Omanska-Klusek A, Wong S, Barrow C, Garcia-Arocena D, Iwahashi C, et al. Evidence of mitochondrial dysfunction in fragile X-associated tremor/ataxia syndrome. Biochem J. 2010;429:545–52.
Napoli E, Song G, Wong S, Hagerman R, Giulivi C. Altered bioenergetics in primary dermal fibroblasts from adult carriers of the FMR1 premutation before the onset of the neurodegenerative disease fragile X-associated tremor/ataxia syndrome. Cerebellum Lond Engl. 2016. doi:10.1007/s12311-016-0779-8
Kiliszek A, Kierzek R, Krzyzosiak WJ, Rypniewski W. Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome. Nucleic Acids Res. 2011;39:7308–15.
Kumar A, Fang P, Park H, Guo M, Nettles KW, Disney MD. A crystal structure of a model of the repeating r(CGG) transcript found in fragile X syndrome. Chembiochem Eur J Chem Biol. 2011;12:2140–2.
Khateb S, Weisman-Shomer P, Hershco-Shani I, Ludwig AL, Fry M. The tetraplex (CGG)n destabilizing proteins hnRNP A2 and CBF-A enhance the in vivo translation of fragile X premutation mRNA. Nucleic Acids Res. 2007;35:5775–88.
Kochetov AV, Prayaga PD, Volkova OA, Sankararamakrishnan R. Hidden coding potential of eukaryotic genomes: nonAUG started ORFs. J Biomol Struct Dyn. 2013;31:103–14.
Plank T-DM, Kieft JS. The structures of nonprotein-coding RNAs that drive internal ribosome entry site function. Wiley Interdiscip Rev RNA. 2012;3:195–212.
Lozano G, Martínez-Salas E. Structural insights into viral IRES-dependent translation mechanisms. Curr Opin Virol. 2015;12:113–20.
Chiang PW, Carpenter LE, Hagerman PJ. The 5′-untranslated region of the FMR1 message facilitates translation by internal ribosome entry. J Biol Chem. 2001;276:37916–21.
Dobson T, Kube E, Timmerman S, Krushel LA. Identifying intrinsic and extrinsic determinants that regulate internal initiation of translation mediated by the FMR1 5′ leader. BMC Mol Biol. 2008;9:89.
Aronin N, DiFiglia M. Huntingtin-lowering strategies in Huntington’s disease: antisense oligonucleotides, small RNAs, and gene editing. Mov Disord Off J Mov Disord Soc. 2014;29:1455–61.
Kordasiewicz HB, Stanek LM, Wancewicz EV, Mazur C, McAlonis MM, Pytel KA, et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–44.
Pretto D, Yrigollen CM, Tang H-T, Williamson J, Espinal G, Iwahashi CK, et al. Clinical and molecular implications of mosaicism in FMR1 full mutations. Front Genet. 2014;5:318.
Park C-Y, Halevy T, Lee DR, Sung JJ, Lee JS, Yanuka O, et al. Reversion of FMR1 methylation and silencing by editing the triplet repeats in fragile X iPSC-derived neurons. Cell Rep. 2015;13:234–41.
Disney MD, Liu B, Yang W-Y, Sellier C, Tran T, Charlet-Berguerand N, et al. A small molecule that targets r(CGG)(exp) and improves defects in fragile X-associated tremor ataxia syndrome. ACS Chem Biol. 2012;7:1711–8.
Tran T, Childs-Disney JL, Liu B, Guan L, Rzuczek S, Disney MD. Targeting the r(CGG) repeats that cause FXTAS with modularly assembled small molecules and oligonucleotides. ACS Chem Biol. 2014;9:904–12.
Tokui K, Adachi H, Waza M, Katsuno M, Minamiyama M, Doi H, et al. 17-DMAG ameliorates polyglutamine-mediated motor neuron degeneration through well-preserved proteasome function in an SBMA model mouse. Hum Mol Genet. 2009;18:898–910.
Waza M, Adachi H, Katsuno M, Minamiyama M, Sang C, Tanaka F, et al. 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat Med. 2005;11:1088–95.
Shi C, Huang X, Zhang B, Zhu D, Luo H, Lu Q, et al. The inhibition of heat shock protein 90 facilitates the degradation of poly-alanine expanded poly (a) binding protein nuclear 1 via the carboxyl terminus of heat shock protein 70-interacting protein. PLoS ONE. 2015;10, e0138936.
Ryno LM, Wiseman RL, Kelly JW. Targeting unfolded protein response signaling pathways to ameliorate protein misfolding diseases. Curr Opin Chem Biol. 2013;17:346–52.
Labbadia J, Cunliffe H, Weiss A, Katsyuba E, Sathasivam K, Seredenina T, et al. Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J Clin Invest. 2011;121:3306–19.
Modi S, Stopeck A, Linden H, Solit D, Chandarlapaty S, Rosen N, et al. HSP90 inhibition is effective in breast cancer: a phase II trial of tanespimycin (17-AAG) plus trastuzumab in patients with HER2-positive metastatic breast cancer progressing on trastuzumab. Clin Cancer Res Off J Am Assoc Cancer Res. 2011;17:5132–9.
Acknowledgments
Our research received funding from the European Union Seventh Framework Programme (FP7/2007-2013), through the European Research Council, under grant agreement RIBOMYLOME_309545 (Gian Gaetano Tartaglia), and from the Fundació La Marató de TV3 (20142731). We also acknowledge support from the Spanish Ministry of Economy and Competitiveness (BFU2011-26206 and BFU2014-55054-P) and “Centro de Excelencia Severo Ochoa 2013–2017” (SEV-2012-0208).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Botta-Orfila, T., Tartaglia, G.G. & Michalon, A. Molecular Pathophysiology of Fragile X-Associated Tremor/Ataxia Syndrome and Perspectives for Drug Development. Cerebellum 15, 599–610 (2016). https://doi.org/10.1007/s12311-016-0800-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12311-016-0800-2