Abstract
Background
Strains of Pediococcus pentosaceus from food and the human gastrointestinal tract have been widely identified, and some have been reported to reduce inflammation, encephalopathy, obesity and fatty liver in animals. In this study, we sequenced the whole genome of P. pentosaceus LI05 (CGMCC 7049), which was isolated from the fecal samples of healthy volunteers, and determined its ability to reduce acute liver injury. No other genomic information for gut-borne P. pentosaceus is currently available in the public domain.
Results
We obtained the draft genome of P. pentosaceus LI05, which was 1,751,578 bp in size and possessed a mean G?+?C content of 37.3%. This genome encoded an abundance of proteins that were protective against acids, bile salts, heat, oxidative stresses, enterocin A, arsenate and universal stresses. Important adhesion proteins were also encoded by the genome. Additionally, P. pentosaceus LI05 genes encoded proteins associated with the biosynthesis of not only three antimicrobials, including prebacteriocin, lysin and colicin V, but also vitamins and functional amino acids, such as riboflavin, folate, biotin, thiamine and gamma-aminobutyrate. A comparison of P. pentosaceus LI05 with all known genomes of food-borne P. pentosaceus strains (ATCC 25745, SL4 and IE-3) revealed that it possessed four novel exopolysaccharide biosynthesis proteins, additional putative environmental stress tolerance proteins and phage-related proteins.
Conclusions
This work demonstrated the probiotic properties of P. pentosaceus LI05 from the gut and the three other food-borne P. pentosaceus strains through genomic analyses. We have revealed the major genomic differences between these strains, providing a framework for understanding the probiotic effects of strain LI05, which exhibits unique physiological and metabolic properties.
Similar content being viewed by others
Background
The genus Pediococcus belongs to the family Lactobacillaceae in the order Lactobacillales. Currently, it is comprised of eleven valid published species, including Pediococcus acidilactici, P. stilesii, P. pentosaceus, P. siamensis, P. cellicola, P. argentinicus, P. parvulus, P. ethanolidurans, P. claussenii, P. inopinatus and P. damnosus[1]. The majority of the members of the genus Pediococcus are used in the food and drink industry as starter and probiotic cultures as well as food spoilers [2]. P. pentosaceus has been intensively investigated and widely employed for food preservation due to its ability to produce antimicrobial agents [3]. Additionally, several strains of P. pentosaceus have been shown to reduce inflammation, encephalopathy [4], obesity and fatty liver [5] in animals. Although food is the main source of P. pentosaceus for humans, the strains of P. pentosaceus adapted to the gastrointestinal tract are dissimilar from those found in food because the former may originate from sub-populations present in food at low numbers that exhibit special adaptive properties [6].
Previously, we have isolated a potential probiotic, P. pentosaceus LI05 (CGMCC 7049), from the fecal samples of healthy volunteers. This strain is tolerant to bile and acid and possesses strong antimicrobial activities against tested enteropathogens. More importantly, the administration of P. pentosaceus LI05 during acute D-galactosamine-induced liver injury in rats was shown to reduce elevated alanine aminotransferase and aspartate aminotransferase levels, prevent the increase of total bilirubin, reduce the histological abnormalities of both the liver and terminal ileum, decrease bacterial translocation, increase the serum levels of IL-10 and result in a cecal microbiome that differ from that of the liver injury control [7].
In this study, we present a summary, classification and the unique characteristics of human gut-borne P. pentosaceus LI05 in addition to a high-quality draft genome sequence and annotations. The probiotic properties of P. pentosaceus LI05 were analyzed using these genomic sequences combined with data from our previous study. Because the genome sequences of P. pentosaceus SL4 from kimchi [8], P. pentosaceus IE-3 from a dairy effluent sample [9], and P. pentosaceus ATCC25745 from plant [10] are now available, this research will provide an essential resource for elucidating the differences between strains isolated from food and the human gastrointestinal tract.
Methods
Determination of cultural, morphological and physiological properties
Growth was investigated under different temperature, pH and NaCl conditions. Cell morphologies, motilities and sporulation activities were examined using transmission electron (H-600, Hitachi Ltd., Tokyo, Japan) microscopy. Phenotypic identification was achieved with API CH50 strips and the API CHL medium system according to the manufacturers instructions (BioMrieux SA, Marcy-lEtoile, France). Other physiological and biochemical tests were conducted as described previously [11]. Phylogenetic analysis was conducted using the neighbor-joining method based on the 16S rRNA and housekeeping gene sequences [12].
Cultural conditions and DNA isolation
After revival using standard methods, the P. pentosaceus LI05 strain (CGMCC 7049) was anaerobically cultured in DeMan-Rogosa-Sharpe (MRS; OXOID, Thermo Fisher Biochemicals Ltd., Beijing, China) broth at 37C for 24 h. Cells were obtained by centrifugation at 8,000 g for 10 min at 4°C. DNA was extracted using the QIAamp DNA Micro Kit according to manufacturer’s guidelines (Qiagen, Westburgb.v., Leusden, The Netherlands).
Genome sequencing and assembly
The genome of P. pentosaceus LI05 was sequenced with the next-generation sequencing platform Illumina HiSeq 2000, and the total number of reads based on a 500-bp library database were 2×11,079,017 (bp). The quality of the sequencing read data was estimated by calculating the quality and GC content of each read. The draft genome sequence was assembled using SOAPdenovo2 [13], and iterative optimization was used to obtain the optimal k-mer value through the use of 31-85 k-mers. The 500-bp libraries were used to build scaffolds, and the SOAPdenovo gap closer software was also used (http://soap.genomics.org.cn/soapdenovo.html). To close the remaining gaps, reference-guided assemblies were carried out with the CLC Genomics Workbench v. 6.05 (CLC bio, Aarbus, Denmark). The combination of de novo assembly and reference-guided assembly was performed manually using the microbial genome-finishing module in the CLC genomics workbench (CLC bio, Aarbus, Denmark). The complete genome sequence of P. pentosaceus ATCC 25745 was used as the reference genome.
Genome annotation
P. pentosaceus LI05 genes were identified using Glimmer [14] together with comparative gene prediction by the direct mapping of the ORFs of the P. pentosaceus ATCC reference strain from the NCBI Genome Database. After a round of manual curation, the unannotated predicted coding sequences (CDS) were translated into amino acid sequences for a query using the NCBI non-redundant database as well as the UniProt, Pfam, COG, and InterPro databases to identify the closest existing homology annotations. Transfer RNA (tRNA) genes were detected using tRNAScanSE [15]. Ribosomal RNAs (rRNAs) were identified using a BLASTn [16] search against the ribosomal RNA databases. Signal peptides were predicted using SignalP 4.0 [17], whereas transmembrane helices in proteins were predicted using TMHMM [18]. The Integrated Microbial Genomes (IMG) platform (http://img.jgi.doe.gov/) was used to support additional gene prediction analyses and manual functional annotations [19].
Comparative genomics
A comparative genomic analysis using BRIG [20] was conducted comparing P. pentosaceus LI05 from the human gastrointestinal tract with three food-borne strains with available genomic sequences, including P. pentosaceus ATCC 25745, SL4 and IE-3. The P. pentosaceu s LI05 genome sequences sharing low identities (<50%) with the other strains were designated as the P. pentosaceus LI05-unique regions. The proteins encoded by the genes that only existed in P. pentosaceus LI05 or that possessed sequence similarities of less than 50% with the three food-borne strains were further analyzed by BLASTp.
Results and discussion
Classification and unique features
P. pentosaceus LI05 is a Gram-negative, non-motile, acid-tolerant, non-sporulating, spherical, facultative anaerobe from the human gastrointestinal tract (Additional file 1: Figure S1). It tolerates 6% NaCl in MRS broth. Growth occurs at 15-45°C and at pH 4-8 but optimally at 37°C. The colonies on the MRS agar were white, smooth, shiny, and circular with complete edges. Some carbohydrates, such as L-arabinose, D-ribose, D-xylose, D-galactose, D-glucose, D-fructose, D-mannose, N-acetylglucosamine, amygdalin, arbutin, salicin, D-cellobiose, D-maltose, D-trehalose, gentiobiose, and D-fucose, can be used as the sole carbon sources, whereas glycerol, erythritol, etc. cannot (Additional file 2: Table S1).
A neighbor-joining tree (Figure 1A) based on the 16S rRNA gene sequence of the strain LI05 shows the phylogenetic relationships between the species of the genus Pediococcus. This organism formed a distinct branch with P. pentosaceus, which was separate from those formed by other members of the genus Pediococcus. Sequence analyses of the dnaA, dnaJ, dnaK, pheS, pryH, recA, recH, tuF, gryB and rplB housekeeping genes were carried out for the definitive identifications of P. pentosaceus LI05, P. pentosaceus ATCC 25745, P. pentosaceus SL4 and P. pentosaceus IE-3. As shown in Figure 1B, the combination of the above housekeeping genes provided good phylogenetic resolution of the four strains. The P. pentosaceus strain IE-3 was the closest evolutionary relative of strain LI05.
The position of P. pentosaceus LI05 relative to the representative strains and the evolutionary relationships of the four strains of P. pentosaceus . A. Phylogenetic tree highlighting the position of P. pentosaceus LI05 relative to the representative strains. The tree was constructed by the neighbor-joining method based on alignments of 16S rRNA gene sequences. Corresponding NCBI accession numbers are shown in parentheses. Numbers at the nodes indicate support values obtained from 1,000 bootstrap replications. B. Phylogenetic tree highlighting the evolutionary relationships of the four strains of P. pentosaceus based on concatenated nucleotide sequences of the dnaA, dnaJ, dnaK, pheS, pryH, recA, recH, tuF, gryB and rplB genes.
Genome properties
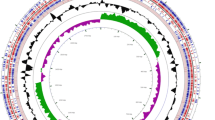
The genome of P. pentosaceus LI05 was sequenced by the Illumina method (see Methods). A total of 11.05 million 100-bp paired-end reads were generated, which provided over 500-fold coverage of the reference genome. High-quality reads with Q?>?30 were assembled using de novo methods to obtain a draft genome of 1.75 Mbp with 8 contigs (the N50 of the assembled contigs was 34.3 Kb; the max length was 318 Kb). The G?+?C content of P. pentosaceus LI05 was 37.29%. For the main chromosome, 1,638 genes were predicted, 1,555 of which were protein-coding genes. A total of 1,321 protein-coding genes were assigned to putative functions, and the remainder were classified as hypothetical proteins. This genome contained 50 tRNAs and a complete 5S-23S-16S rRNA gene family. The properties and statistics of the genome are shown in Table 1 and Figure 2. As shown in Figure 3, the genome sequence of P. pentosaceus LI05 was highly conserved compared with those of P. pentosaceus ATCC 25745, P. pentosaceus SL4 and P. pentosaceus IE-3.
Genome of P. pentosaceus LI05 exhibits probiotic properties
In a previous study, we have observed that P. pentosaceus LI05 is resistant to gastric acidity and bile compounds [7]. This was supported by the genomic data from this study, in which a gene encoding cholylglycine hydrolase, which is related to bile salt resistance, and genes encoding F0F1 ATP synthases, which are important for acid tolerance, were detected (Table 2). Additionally, six universal stress proteins (UspA), the chaperone protein DnaJ, the cofactor GrpE, which participates in the hyperosmotic and heat shock responses, the chaperone GroEL, which protects against environmental stresses, an enterocin A immunity family protein, an arsenate reductase, and methionine sulfoxide reductase A, which protects against oxidative stresses, were annotated. These representative stress resistance genes were highly conserved between P. pentosaceus LI05 and P. pentosaceus ATCC 25745, P. pentosaceus IE-3 or P. pentosaceus sL4, but most of them showed significant divergences from other species.
The ability to adhere to gastrointestinal mucosa is an important property of most probiotics [21],[22]. Several proteins encoded by P. pentosaceus LI05 genes had predicted adhesive potentials (Table 3). For example, sortase attaches surface proteins, including enzymes, pilins and adhesion-mediating large surface glycoproteins, to cell walls. Other proteins included a pilin-like competence protein ComGC, elongation factor Tu (EF-Tu), an enolase capable of binding to host extracellular fibronectin and the pilus biosynthesis protein HicB. Abundant adhesion proteins encoded by the genomic regions were consistent with the strong adhesion properties of P. pentosaceus LI05. However, these proteins have also been predicted in the other tested strains. These findings may represent a possible reason for the extensive colonization of P. pentosaceus in the gut. However, the examinations of many more genes or proteins may be required to evaluate the adhesive abilities of probiotics.
The P. pentosaceus LI05 genes also encoded three antimicrobials, which is consistent with the excellent antimicrobial ability of this strain. As shown in Table 3, genes encoding prebacteriocin were annotated in the genomes of both P. pentosaceus LI05 and P. pentosaceus ATCC 25745. Alternatively, the pedA gene (PCPN_1274) encoding pediocin PA-1 was detected in P. pentosaceus IE-3, but it was not identical to the prebacteriocin gene of P. pentosaceus LI05. Furthermore, genes encoding colicin V, which is a peptide antibiotic that kills sensitive cells by disrupting their membrane potentials [23], were found in these four P. pentosaceus strains. However, the colicin V discovered in strain L105 was different from that of the other spicies. Additionally, genes encoding lysin were detected in P. pentosaceus LI05 and P. pentosaceus ATCC 25745. As an antimicrobial agent, lysin is potentially immunogenic [24]. Therefore, P. pentosaceus LI05 can achieve “competitive exclusion” not only by limiting the surface area available but also by secreting antimicrobial substances.
In the genome of P. pentosaceus LI05, we also detected potentially beneficial properties that were not experimentally confirmed. This strain contained genes involved in the biosynthesis of not only important vitamins, such as riboflavin, folate, thiamine and biotin but also of functional factors, such as gamma-aminobutyrate (Table 3) [25]. In Gram-positive bacteria, peptidoglycan is one of the most important host immune regulators [26]. Although the genes and coding proteins related to the peptidoglycan pathway were conserved in the four strains of P. pentosaceus, they were not significantly similar to those of the other species. These findings will contribute to the elucidation of the mechanisms of immune regulation in P. pentosaceus LI05.
Comparisons with other fully sequenced genomes
Fifty-three proteins encoded by P. pentosaceus LI05 genes were not detected or had sequence similarities of less than 50% in the comparative analysis with the three known food-borne strains, P. pentosaceus ATCC 25745, SL4 and IE-3. Among these proteins, 21 hypothetical proteins with no clear functions were not further analyzed; the other 32 proteins are listed in Table 4, demonstrated in Figure 3, and further discussed below.
Five putative exopolysaccharide biosynthesis proteins were detected only in P. pentosaceus LI05, including an epimerase, a capsular polysaccharide biosynthesis protein, two glycosyltransferases (key enzymes for the biosyntheses of the exopolysaccharide repeating units) and a polysaccharide biosynthesis protein. Four of these enzymes need to be examined in further detail because they are not only potentially novel but also probably induce variations in the structures of their encoded polysaccharides that may have influenced adherence, biofilm formation and the nature of the immune response [27].
P. pentosaceus LI05 was characterized by three extra-environmental stress tolerance proteins, including a putative ferritin-like DNA-binding protein, which maintains a steady state of iron ions and responds to stresses, such as those involving temperature, humidity, and ionizing and redox processes [28], a putative PadR family transcriptional regulator, which functions against phenolic acid stress, and a putative ThiJ/PfpI family protein, which is involved in cellular protection against environmental stresses [29].
Fourteen proteins related to the intrusion of exogenous DNA were identified in P. pentosaceus LI05. One group was comprised of twelve prophage-related proteins, including a phage integrase family site-specific recombinase, two integrases, a putative prophage repressor, two phage proteins, a replisome organizer, a terminase, two minor capsid proteins, a capsid protein and a tail protein. It is not rare for bacteria to contain multiple prophages in their chromosomes, which then constitute a sizable proportion of their total chromosomal material [30]. Pathogenic, commensal, and symbiotic bacteria have been observed to play roles in a variety of bacterial adaptations in hosts [31]. Phage-related proteins were encoded by genes in each of the three food-borne strains. The other genes detected in the P. pentosaceus LI05 included two encoding bacterial DNA type I restriction endonucleases, which are involved in prokaryotic DNA restriction-modification mechanisms that protect the bacteria against invading foreign DNA [32].
Two putative doxorubicin-daunorubicin resistance proteins existed in P. pentosaceus LI05. One ORF encoded DrrA, which is part of the ABC transporter complex DrrAB. The other ORF encoded DrrC, which is part of the ABC transporter permease protein. This finding partially reflects the complex interactions between drugs and gut-associated microbes [33]. Both daunorubicin and doxorubicin are antitumor drugs and are thus not suitable for antibacterial applications. Therefore, these two genes will not affect the control of P. pentosaceus LI05.
Additionally, there were eight extra putative multifunctional proteins in P. pentosaceus LI05. These included a TetR family transcriptional regulator, an ABC transporter permease, an exonuclease ABC subunit A, a transposase, an acetyl xylan esterase, a PadR family transcriptional regulator, a membrane protein and a TraX family protein.
Conclusions
Strains of P. pentosaceus are frequently identified in food and in the human gastrointestinal tract and are known to reduce inflammation, encephalopathy, obesity and fatty liver in animals. Therefore, it is imperative to study the probiotic ability of this organism. Future studies will focus on delineating the interactions between the host and P. pentosaceus. The genome sequences of P. pentosaceus LI05 isolated from the human gastrointestinal tract allow for a deeper understanding of its probiotic abilities, facilitating the future development of drugs for microbiota-related diseases.
Availability of supporting data
The whole-genome sequencing project of P. pentosaceus LI05 has been submitted to GenBank under the project accession number PRJNA237570. The project version entailing the draft assembly described herein has been deposited under the accession number JDVW00000000.
Authors’ contributions
L-JL designed the study, interpreted the results and edited the manuscript. L-XL and Y-DL conducted the Illumina sequencing, performed the assemblies, analyzed the genome, and performed the annotations. X-JH provided advice related to the outbreak and strain features, characterized the strain and maintained it in pure cultures. H-YS contributed to the microbiology of the strain and prepared high-molecular-weight DNA for the genome sequencing. All authors read and approved the manuscript prior to submission.
Additional files
References
Wieme A, Cleenwerck I, Van Landschoot A, Vandamme P:Pediococcus lolii DSM 19927 T and JCM 15055 T are strains of Pediococcus acidilactici. Int J Syst Evol Microbiol. 2012, 62: 3105-3108. 10.1099/ijs.0.046201-0.
Leroy F, Verluyten J, De Vuyst L: Functional meat starter cultures for improved sausage fermentation. Int J Food Microbiol. 2006, 106: 270-285. 10.1016/j.ijfoodmicro.2005.06.027.
Martino ME, Maifreni M, Marino M, Bartolomeoli I, Carraro L, Fasolato L, Cardazzo B: Genotypic and phenotypic diversity of Pediococcus pentosaceus strains isolated from food matrices and characterisation of the penocin operon. Antonie Van Leeuwenhoek. 2013, 103: 1149-1163. 10.1007/s10482-013-9897-1.
Bengmark S: Bio-ecological control of chronic liver disease and encephalopathy. Metab Brain Dis. 2009, 24: 223-236. 10.1007/s11011-008-9128-z.
Zhao X, Higashikawa F, Noda M, Kawamura Y, Matoba Y, Kumagai T, Sugiyama M: The obesity and fatty liver are reduced by plant-derived Pediococcus pentosaceus LP28 in high fat diet-induced obese mice. PLoS One. 2012, 7: e30696-10.1371/journal.pone.0030696.
Varsha KK, Priya S, Devendra L, Nampoothiri KM: Control of Spoilage Fungi by Protective Lactic Acid Bacteria Displaying Probiotic Properties. Appl Biochem Biotechnol. 2014, 172: 3402-3413. 10.1007/s12010-014-0779-4.
Lv LX, Hu XJ, Qian GR, Zhang H, Lu HF, Zheng BW, Jiang L, Li LJ: Administration of Lactobacillus salivarius LI01 or Pediococcus pentosaceus LI05 improves acute liver injury induced by D-galactosamine in rats. Appl Microbiol Biotechnol. 2014, 98: 5619-5632. 10.1007/s00253-014-5638-2.
Dantoft SH, Bielak EM, Seo JG, Chung MJ, Jensen PR: Complete genome sequence of Pediococcus pentosaceus strain SL4. Genome Announc. 2013, 26: e01106-e01113.
Midha S, Ranjan M, Sharma V, Kumari A, Singh PK, Korpole S, Patil PB: Genome sequence of Pediococcus pentosaceus strain IE-3. J Bacteriol. 2012, 194: 4468-10.1128/JB.00897-12.
Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Díaz-Muñiz I, Dosti B, Smeianov V, Wechter W, Barabote R: Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A. 2006, 103: 15611-15616. 10.1073/pnas.0607117103.
Smibert RM, Krieg NR: Phenotypic Characterization. Methods for General and Molecular Bacteriology. 1994, American Society for Microbiology Press, Washington, DC, USA, 607-654.
Tamura K, Dudley J, Nei M, Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007, 24: 1596-1599. 10.1093/molbev/msm092.
Luo R, Liu BH, Xie YL: SOAPdenovo2: an empirically improved memory-efficient short-read de novoassembler. GigaScience. 2012, 1: 18-10.1186/2047-217X-1-18.
Delcher AL, Bratke KA, Powers EC, Salzberg SL: Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007, 23: 673-679. 10.1093/bioinformatics/btm009.
Lowe TM, Eddy SR: tRNAscan-SE: a program for improved detection of transfer RNAgenes in genomic sequence. Nucleic Acids Res. 1997, 25: 955-964. 10.1093/nar/25.5.0955.
Benson G: Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999, 27: 573-580. 10.1093/nar/27.2.573.
Petersen TN, Brunak S, von Heijne G, Nielsen H: SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011, 8: 785-786. 10.1038/nmeth.1701.
Krogh A, Larsson B, von Heijne G, Sonnhammer EL: Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001, 305: 567-580. 10.1006/jmbi.2000.4315.
Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Pillay M, Ratner A, Huang J, Woyke T, Huntemann M, Anderson I, Billis K, Varghese N, Mavromatis K, Pati A, Ivanova NN, Kyrpides NC: IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014, 42: D560-D567. 10.1093/nar/gkt963.
Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA: BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011, 12: 402-10.1186/1471-2164-12-402.
Senan S, Prajapati JB, Joshi CG: Whole-genome based validation of the adaptive properties of Indian origin probiotic Lactobacillus helveticus MTCC 5463. J Sci Food Agric. 2014,doi:10.1002/jsfa.6721.
Grover S, Rashmi HM, Srivastava AK, Batish VK: Probiotics for human health -new innovations and emerging trends. Gut Pathog. 2012, 4: 15-10.1186/1757-4749-4-15.
Kim YC, Tarr AW, Penfold CN: Colicin import into E. coli cells: a model system for insights into the import mechanisms of bacteriocins. Biochim Biophys Acta. 1843, 2014: 1717-1731.
Pastagia M, Schuch R, Fischetti VA, Huang DB: Lysins: the arrival of pathogen-directed anti-infectives. J Med Microbiol. 2013, 62: 1506-1516. 10.1099/jmm.0.061028-0.
Capozzi V, Russo P, Duenas MT, Lopez P, Spano G: Lactic acid bacteria producing B-group vitamins: a great potential for functional cereals products. Appl Microbiol Biotechnol. 2012, 96: 1383-1394. 10.1007/s00253-012-4440-2.
Kanmani P, Satish Kumar R, Yuvaraj N, Paari KA, Pattukumar V, Arul V: Probiotics and its functionally valuable products-a review. Crit Rev Food Sci Nutr. 2013, 53: 641-658. 10.1080/10408398.2011.553752.
Nadkarni MA, Chen Z, Wilkins MR, Hunter N: Comparative genome analysis of Lactobacillus rhamnosus clinical isolates from initial stages of dental pulp infection: identification of a new exopolysaccharide cluster. PLoS One. 2014, 9: e90643-10.1371/journal.pone.0090643.
Smith JL: The physiological role of ferritin-like compounds in bacteria. Crit Rev Microbiol. 2004, 30: 173-185. 10.1080/10408410490435151.
Zhan D, Han W, Feng Y: Experimental and computational studies indicate the mutation of Glu12 to increase the thermostability of oligomeric protease from Pyrococcus horikoshii. J Mol Model. 2011, 17: 1241-1249. 10.1007/s00894-010-0819-0.
Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H: Prophage genomics. Microbiol Mol Biol Rev. 2003, 67: 238-276. 10.1128/MMBR.67.2.238-276.2003.
Hooper LV, Gordon JI: Commensal host-bacterial relationships in the gut. Science. 2001, 292: 1115-1118. 10.1126/science.1058709.
Loenen WA, Raleigh EA: The other face of restriction: modification-dependent enzymes. Nucleic Acids Res. 2014, 42: 56-69. 10.1093/nar/gkt747.
Saad R, Rizkallah MR, Aziz RK: Gut Pharmacomicrobiomics: the tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut Pathog. 2012, 4: 16-10.1186/1757-4749-4-16.
Acknowledgments
This study was supported by the National Basic Research Program of China (973 Program) (No. 2013CB531401) and the Key Program of the National Natural Science Foundation of China (No. 81330011).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare no competing interests.
Electronic supplementary material
13099_2014_36_MOESM1_ESM.pdf
Additional file 1: Figure S1.: Scanning electron micrograph (A) and transmission electron micrograph (B) of P. pentosaceus LI05. (PDF 517 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Lv, LX., Li, YD., Hu, XJ. et al. Whole-genome sequence assembly of Pediococcus pentosaceus LI05 (CGMCC 7049) from the human gastrointestinal tract and comparative analysis with representative sequences from three food-borne strains. Gut Pathog 6, 36 (2014). https://doi.org/10.1186/s13099-014-0036-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13099-014-0036-y