Abstract
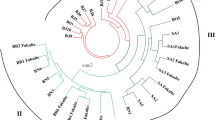
Molecular markers provide facilities in order to study genetic diversity and relationship among genotypes. In this study, genetic diversity among 35 genotype of Brassica sp. (belonging B. napus, B. juncea, B. rapa, B. nigra) were determined using 13 ISSR, 3 IRAP markers and 18 REMAP (primer combinations of ISSR and retrotransposon primer). The percentage of polymorphism for ISSR, IRAP and REMAP was 96.38, 94 and 96%, respectively. By comparison between markers, ISSRs indicated the highest expected heterozygosity (He) and Shannon’s information index (I) with value of 0.34 and 0.51, respectively, while REMAP marker had by far the highest number of polymorphic bands (340) and marker index (7.1) among all fragments scored over all markers. In pattern of clustering based on Bayesian methods, K = 8 was resulted for combined data clustering that was more organized clustering for genotypes compared to others. This research suggests the combined data of ISSR, IRAP and REMAP markers are most reliable than each solely marker whilst have been clustered genotypes in their taxonomic classification of Brassica without any mixture. Principle coordinate analysis (PCoA) separated 35 genotypes in four groups which all of genotypes were clustered correctly based on their taxonomic classification. The findings of this study provide the valuable insight into the Brassica species relationships in terms of similarity among genotypes which can be helpful in breeding programs, and also demonstrate that retrotransposon markers are legible for genetic diversity and next genetic analysis in Brassica genus.
Similar content being viewed by others
References
Food and Agriculture Organization (FAO), Crop Production Statistics, 2012. http://www.fao.org.
Fu, T.D., Yang, G.S., Tu, J.X. and Ma, C.Z., The present and future of rapeseed production in China, China Oils Fats, 2003, vol. 28, pp.11–13.
McVetty, P.B.E., Scarth, R., Fernando, W.G.D., et al., Brassica seed quality breeding at the university of Manitoba, in Proceedings of 12th International Rapeseed Congress: Genetics and Breeding, Wuhan, China, 2007, pp. 2–4.
Knapp, S.J., Marker-assisted selection as a strategy for increasing the probability of selecting superior genotypes, Crop. Sci., 1998, vol. 38, no. 5, pp. 1164–1174.
Curn, V. and Zaludova, J., Fingerprinting of oilseed rape cultivars, Adv. Bot. Res., 2007, vol. 45, pp. 155–179.
Pradeep Reddy, M., Sarla, N., and Siddiq, E.A., Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding, Euphytica, 2002, vol. 128, no. 1, pp. 9–17.
Joshi, S.P., Gupta, V.S., Aggarwal, R.K., et al., Genetic diversity and phylogenetic relationship as revealed by inter-simple sequence repeat (ISSR) polymorphism in the genus Oryza, Theor. Appl. Genet., 2000, vol. 100, no. 8, pp. 1311–1320.
Hajiyev, E.S., Akparov, Z.I., Aliyev, R.T., et al., Genetic polymorphism of durum wheat (Triticum durum Desf.) accessions of Azerbaijan, Russ. J. Genet., 2015, vol. 51, no. 9, pp. 863–870.
Liu, B. and Wendel, J.F., Inter simple sequence repeat (ISSR) polymorphisms as a genetic marker system in cotton, Mol. Ecol. Notes, 2001, vol.1, no. 3, pp. 205–208.
Wakui, K., Iwata, H., Takahashi, Y., and Fujigaki, J., Assessment of the congruity of genetic relationships and variation revealed by individual-and bulked-samples-based approaches using RAPD and ISSR markers in Japanese turnip (Brassica rapa ssp. rapa) cultivars, Breed. Sci., 2009, vol. 59, no. 4, pp. 447–452.
Parsaeian, M., Mirlohi, A., Saeidi, G., Study of genetic variation in sesame (Sesamum indicum L.) using agromorphological traits and ISSR markers, Russ. J. Genet., 2011, vol. 47, no. 3, pp. 314–321.
Grzebelus, D., Transposon insertion polymorphism as a new source of molecular markers, J. Fruit Ornam. Plant Res., 2006, vol. 14, no. 1, pp. 21–29.
Lee, D., Ellis, T.H.N., Turner, L., et al., A copia-like element in Pisum demonstrates the uses of dispersed repeated sequences in genetic analysis, Plant Mol. Biol., 1990, vol.15, no. 5, pp. 707–722.
Kumar, A., The adventures of the Ty1-copia group of retrotransposons in plants, Trends Genet., 1996, vol. 12, no. 2, p. 4143.
Li, M.T., Chen, X., and Meng, J.L., Intersubgenomic heterosis in rapeseed production with a partial newtyped Brassica napus containing subgenome Ar from B. rapa and Cc from Brassica carinata, Crop Sci., 2006, vol. 46, no. 1, pp. 234–242.
SanMiguel, P. and Bennetzen, J.I., Evidence that a recent increase in maize genome size was caused by the massive amplification of intergene retrotransposons, Ann. Bot., 1998, vol. 81, pp. 37–44.
Schulman, A.H., Flavell, A.J., and Ellis, T.H., The application of LTR retrotransposons as genetic markers in plants, Methods Mol. Biol., 2004, vol. 260, pp. 145–173.
Waugh, R., McLean, K., Flavell, A.J., et al., Genetic distribution of BARE-1 retrotransposable elements in the barley genome revealed by sequence-specific amplification polymorphisms (S-SAP), Mol. Gen. Genet., 1997, vol. 253, no. 6, pp. 687–694.
Kalendar, R., Grob, T., Regina, M., et al., IRAP and REMAP: two retrotransposon-based DNA fingerprinting techniques, Theor. Appl. Genet., 1999, vol. 98, no. 5, pp. 704–711.
Kalendar R. and Schulman, A.H., IRAP and REMAP for retrotransposon-based genotyping and fingerprinting, Nat. Protocl., 2006, vol. 1, no. 5, pp. 2478–2484.
Saeidi, H., Rahiminejad, M.R., and Heslop-Harrison, J.S., Retroelement insertional polymorphisms, diversity and phylogeography within diploid, D-genome Aegilops tauschii (Triticeae, Poaceae) sub-taxa in Iran, Ann. Bot., 2008, vol. 101, no. 6, pp. 855–861.
Manninen, O., Kalendar, R., Robinson, J., and Schulman, A.H., Application of BARE-1 retrotransposon markers to the mapping of a major resistance gene for net blotch in barley, Mol. Gen. Genet., 2000, vol. 264, no. 3, pp. 325–334.
Leigh, F., Kalendar, R., Lea, V., et al., Comparison of the utility of barley retrotransposon families for genetic analysis by molecular marker techniques, Mol. Gen. Genomics, 2003, vol. 269, no. 4, pp. 464–474.
Bernet, G.P., Mestre, P.F., Pina, J.A., and Asins, M.J., Molecular discrimination of lemon cultivars, Hortscience, 2004, vol. 39, no. 1, pp. 165–169.
Alavi-Kia, S.S., Mohammadi, S.A., Aharizad, S., and Moghaddam, M., Analysis of genetic diversity and phylogenetic relationships in Crocus genus of Iran using inter-retrotransposon amplified polymorphism, Biotechnol. Biotec. EQ, 2008, vol. 22, no. 3, pp. 795–800.
Yuying, S., Xiajun, D., Fei, W., et al., Analysis of genetic diversity in Japanese apricot (Prunus mume Sieb. et Zucc.) based on REMAP and IRAP molecular markers, Sci. Hortic., 2011, vol. 132, no. 1, pp. 50–58.
Huo, H., Conner, J.A., and Ozias-Akins, P., Genetic mapping of the apospory-specific genomic region in Pennisetum squamulatum using retrotransposon-based molecular markers, Theor. Appl. Genet., 2009, vol. 119, no. 2, pp. 199–212.
Boronnikova, S.V. and Kalendar, R.N., Using IRAP markers for analysis of genetic variability in populations of resource and rare species of plants, Russ. J. Genet., 2010, vol. 46, no. 1, pp. 36–42.
Cakmak, B., Marakli, S., and Gozukirmizi, N., SIRE1 retrotransposons in barley (Hordeum vulgare L.), Russ. J. Genet., 2015, vol. 51, no. 7, pp. 661–672.
Cabo, S., Carvalho, A., Rocha, L., and Lima-Brito, J., IRAP, REMAP and ISSR fingerprinting in newly formed hexaploid Tritordeum (X Tritordeum Ascherson et Graebner) and respective parental species, Plant Mol. Biol. Rep., 2013, vol. 32, no. 3, pp. 761–770.
Carvalho, A., Guedes-Pinto, H., Martins-Lopes, P., and Lima-Brito, J., Genetic variability analysis in Old Portuguese bread wheat cultivars assayed by IRAP and REMAP markers, Ann. Appl. Biol., 2010, vol. 156, no. 3, pp. 337–345.
Carvalho, A., Guedes-Pinto, H., and Lima-Brito, J., Genetic diversity in Old Portuguese durum wheat cultivars assessed by retrotransposon based markers, Plant Mol. Biol. Rep., 2012, vol. 30, no. 3, pp. 578–589.
Dellaporta, S.L., Wood J., and Tickes, J.B., A plant molecular DNA mini-preparation, Version II, Plant Mol. Biol. Rep., 1993, vol. 1, no. 4, pp. 19–21.
Kalendar, R., Tanskanen, J., Chang, W., et al., Cassandra retrotransposons carry independently transcribed 5S RNA, Proc. Natl. Acad. Sci. U.S.A., 2007, vol. 105, no. 15, pp. 5833–5838.
Peakall, R. and Smouse, P.E., GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update, Bioinformatics, 2012, vol. 28, no. 19, pp. 2537–2539.
Pritchard, J.K., Wen, W., and Falush, D., Documentation for Structure software v. 2.3, 2010.
Evanno, G., Regnaut, S., and Goudet, J., Detecting the number of clusters of individuals using the software Structure: a simulation study, Mol. Ecol., 2005, vol. 14, no. 8, pp. 2611–2620.
Huson, D.H. and Bryant, D., Documentation for SplitsTree4 v. 4.12.6, 2012.
Scariot, V., De Keyser, E., Handa, T., and De Riek, J., Comparative study of the discriminating capacity and effectiveness of AFLP, STMS and EST markers in assessing genetic relationships among evergreen azaleas, Plant Breed., 2007, vol. 126, no. 2, pp. 207–212.
Powell, W., Morgante, M., Andre, C., et al., The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis, Mol. Breed., 1996, vol. 2, no. 3, pp. 225–238.
Branco, C.J., Vieira, E.A., Malone, G., et al., IRAP and REMAP assessments of genetic similarity in rice, J. Appl. Genet., 2007, vol. 48, no. 2, pp. 107–113.
Abdollahi Mandoulakani, B., Piri, Y., Darvishzadeh, R., et al., Retroelement insertional polymorphism and genetic diversity in Medicago sativa populations revealed by IRAP and REMAP markers, Plant Mol. Biol. Rep., 2012, vol. 30, no. 2. pp. 286–296.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Mahjoob, B., Zarini, H.N., Hashemi, S.H. et al. Comparison of ISSR, IRAP and REMAP markers for assessing genetic diversity in different species of Brassica sp.. Russ J Genet 52, 1272–1281 (2016). https://doi.org/10.1134/S1022795416120073
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795416120073