Abstract
A major challenge in therapeutic approaches applying hematopoietic stem cells (HSCs) is the cell quantity. The primary objective of this study was to predict the miRNAs and anti-miRNAs using bioinformatics tools and investigate their effects on the expression levels of key genes predicted in the improvement of proliferation, and the inhibition of differentiation in HSCs isolated from Human umbilical cord blood (HUCB). A network including genes related to the differentiation and proliferation stages of HSCs was constructed by enriching data of text (PubMed) and StemChecker server with KEGG signaling pathways, and was improved using GEO datasets. Bioinformatics tools predicted a profile from miRNAs containing miR-20a-5p, miR-423-5p, and chimeric anti-miRNA constructed from 5′-miR-340/3′-miR-524 for the high-score genes (RB1, SMAD4, STAT1, CALML4, GNG13, and CDKN1A/CDKN1B genes) in the network. The miRNAs and anti-miRNA were transferred into HSCs using polyethylenimine (PEI). The gene expression levels were estimated using the RT-qPCR technique in the PEI + (miRNA/anti-miRNA)-contained cell groups (n = 6). Furthermore, CD markers (90, 16, and 45) were evaluated using flow cytometry. Strong relationships were found between the high-score genes, miRNAs, and chimeric anti-miRNA. The RB1, SMAD4, and STAT1 gene expression levels were decreased by miR-20a-5p (P < 0.05). Additionally, the anti-miRNA increased the gene expression level of GNG13 (P < 0.05), whereas the miR-423-5p decreased the CDKN1A gene expression level (P < 0.01). The cellular count also increased significantly (P < 0.05) but the CD45 differentiation marker did not change in the cell groups. The study revealed the predicted miRNA/anti-miRNA profile expands HSCs isolated from HUCB. While miR-20a-5p suppressed the RB1, SMAD4, and STAT1 genes involved in cellular differentiation, the anti-miRNA promoted the GNG13 gene related to the proliferation process. Notably, the mixed miRNA/anti-miRNA group exhibited the highest cellular expansion. This approach could hold promise for enhancing the cell quantity in HSC therapy.
Similar content being viewed by others
Introduction
It is well known that stem cells can proliferate and give rise to new differentiated cells. Hematopoietic stem cells (HSCs) are a specific class that preserves blood cell balance and are found in various sources, such as umbilical cord blood, bone marrow, and peripheral blood1. These cells can differentiate into other blood cells and may be used in the treatment of various blood disorders2. However, obtaining an applicable count of cells before they differentiate is a significant challenge for therapeutic goals3.
Some studies have focused on the gene profiles and signaling pathways involved in the differentiation and proliferation of HSCs4. A suitable population of HSCs may be produced by identifying the factors and using complementary laboratory-based therapies that control the cell cycle and differentiation processes in HSCs. According to some reports, the regulation of these processes in HSCs is greatly related to signaling pathways including WNT, NOTCH, TGFB, FGF, RAS, and JACK-STAT5,6. Furthermore, some genes such as Bmi-1, Nanog, Oct4, Sox2, HOXB4, and Ezh2 are reported as master regulators of differentiation and proliferation processes in the HSCs7,8.
By employing bioinformatics tools to analyze high-throughput data, the gene profiles might be used to find the signaling pathways involved in various cellular processes and be enriched to understand the gene functions and their regulatory factors, including miRNAs9,10. It is well known that the function of miRNAs may be controlled by other molecules, such as anti-miRNAs, resulting in modification of the expression of target genes11. Thus, a proper design for the simultaneous use of miRNAs and anti-miRNAs can affect the expression levels of genes involved in the proliferation and differentiation of HSCs.
The aim of this study was to predict the genes, miRNAs, and anti-miRNAs involved in the differentiation and proliferation processes of HSCs isolated from HUCB using bioinformatics tools. Then, the predicted miRNA/anti-miRNA profile was studied on the high-score gene expression levels to improve the expansion of HSCs.
Materials and methods
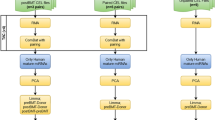
Gene and miRNA prediction
Data mining
The data were collected from publications (PubMed, 2018–2024) with the keywords containing ((“Hematopoietic stem cells” OR “Blood stem cells” OR “Hematopoiesis” OR “Hematopoietic cell development” OR “Hematopoietic cell replication” OR “Hematopoietic stem cell self-renewal” OR “Hematopoietic stem cell differentiation” OR “Hematopoietic stem cell expansion” OR “Hematopoietic stem cell niche” OR “Hematopoietic stem cell transplantation”) AND (“gene regulation” OR “cell proliferation” OR “gene expression” OR “gene function” OR “hematopoietic genes” OR “gene pathways” OR “gene networks” OR “gene targets” OR “gene interactions” OR “gene therapy”)). The focus of the data collection was on identifying genes associated with their crucial roles in controlling the proliferation and differentiation processes of HSCs3,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28. Furthermore, the genes were extracted and annotated from the high-throughput studies on HSCs in the StemChecker web server (http://stemchecker.sysbiolab.eu/)29. The StemChecker database is specialized in curating research on stem cell types. It compiles high-throughput studies such as RNA sequencing and microarrays that have identified critical genes for stem cell growth and differentiation. Then, the gene collection was enriched using the KEGG database (www.kegg.jp/kegg/kegg1.html) and the Enrichr database (https://maayanlab.cloud/Enrichr/) to identify the signaling pathways that influence the various functions of HSCs30 (Fig. 1A). These databases serve as a rich resource for deciphering cellular signaling pathways. By incorporating biological parameters, the databases can pinpoint pathways associated with a specific set of genes. Finally, the genes involved in the signaling pathways were the subjects to construct the gene network.
Gene network
A primary gene network was constructed using STRING (https://string-db.org/, interaction score > 0.9, cluster > 5 nodes) (Fig. 1B) from the genes of significant signaling pathways. Then, the gene network was transported into Cytoscape software (version 3.9.1)31 to visualize and improve the edges and nodes. The STRING creates a network that shows the communicational edges between genes. Furthermore, Cytoscape facilitates network manipulation and determines statistical parameters to further analyze the network during the subsequent stages.
Improvement of the gene network using GEO data
The gene fold data obtained from the GEO (https://www.ncbi.nlm.nih.gov/gds/?term =) datasets (GSE107497, GSE125345, and GSE179928) were used to improve the gene network (Table 1) (Fig. 1C). The datasets were analyzed with the GEO2R and R programming tools using the specialized Limma and DESeq2 packages to merge and normalize the data. The genes exhibiting significant up and low expression levels (P-value < 0.05 and |Log Fold Change (LFC)|> 1) in the cellular proliferation stage were compared to the differentiation process (Table 2) so that the intersection of genes were defined two sets. (1) The high and low expression genes (n = 108) predicted in the proliferation and self-renewal of HSCs. (2) The high and low expression genes (n = 103) in the differentiation stage of HSCs. The nodes on the gene network were improved using the absolute values of gene fold data and displayed the proliferation and differentiation clusters.
Gene‑miRNA bipartite relationship
To pinpoint miRNAs that potentially regulate the high-score nodes (high-size nodes) in HSC proliferation and differentiation stages, the miRTarBase, DIANA-TarBase, and ENCORI/starBase databases were applied. Each database employs distinct algorithms and experimental data to predict miRNA-target gene interactions. The scores in each database reflect the strength of prediction or confidence of binding miRNA to the target gene. Higher scores typically indicate a more likely interaction (Fig. 1D). The final grading of the gene-miRNA bipartite was obtained from the sum of database scores and was represented as edge thickness.
Cellular isolation and culture
The HUCB bags (25 ml, citrate, phosphate, and dextrose) containing the suitable HSC counts were evaluated and obtained by the Iranian Blood Transfusion Organization (IBTO) in conformity with the community ethical committee’s ethical guidelines (IR.IUMS.FMD.REC.1402.048). Each UCB unit was diluted 1:1 with phosphate buffered solution and transferred into a 50 ml tube containing 15 ml Ficoll Hypaque solution (Amersham Pharmacia, Piscataway, NJ; density, 1.077 g/mL). Mononuclear cells (MNCs) were obtained from the interphase layer (above the Ficoll layer) after centrifuging at 2800 rpm (25 min) (Fig. 1E). MNCs were then washed twice with DMEM F12 (10% FBS, Gibco, Thermo Fisher Scientific-US), re-suspended, and seeded in DMEM F12 media (20% FBS, 1% Pen-Strep (Sigma-Aldrich Co., St Louis, MO, USA)) in humidified conditions (37 °C, 5% CO2). Some studies reported that SCF, TPO and other growth factors may trigger the differentiation process32,33. To diminish these effects, the medium was limited with these factors and supplemented with 150 µg/ml apo-transferrin34, 50 µg/ml insulin, 20 units/ml Erythropoietin (EPO), 50 ng/ml growth hormone (GH)35,36, vitamin A and E each 50 µM, nicotinamide 10 mM,37,38,39 and non-essential amino acids (NEAA)40 (Gibco, cat No. 11140–035). The non-adherent cells including HSCs, were isolated during several passages41. In each passage, the adherent cells were discarded, and the supernatant cells were used only for the next passage.
Experimental study
Cell groups
The gene fold data of GSE179928 obtained in 4-day period were used to improve the nodes on the gene network. However, to better observe the variance changes of genes and the self-renewal and differentiation CD markers that may appear due to the time-dependent cell phenotype events, the pre-interventional culture period of HSCs increased up to 6 days as the longer periods were reported in other studies35,42. On the sixth day, the cultured (enriched DMEM F12) cells (1 × 106) were divided into six groups, including the polyethyleneimine (PEI) (25 kDa), PEI + miR Synthetic (non-functional nucleotide sequence), PEI + miR-20a-5p, PEI + miR-423-5p, PEI + anti-miRNA, and PEI + miR Synthetic + miR-20a-5p + miR-423-5p + and anti-miRNA groups (Fig. 1F).
Transfer of miRNA/anti-miRNA into HSCs
The anti-miRNA was designed by connecting 11 reverse complement bases of 5′miR-340-5p (MIMAT0004692: 5′-UUAUAAAGCAAUGAGACUGAUU-3′) and 11 reverse complement bases of 3′miR-524-5p (MIMAT0002849: 5′-CUACAAAGGGAAGCACUUUCUC-3′), which were separated from each other with AT nucleotide bases so that the chimeric construct might bind to both the miRNAs (Table 3). PEI (Sigma Aldrich, Cat. No. 408,727) was used to transfer the miRNA/anti-miRNA into HSCs. A solution of PEI was prepared by shaking at 37 °C (20 mg/ml DEPC water). Also, a solution of miR-20a-5p (MIMAT0000075), miR-423-5p (MIMAT0004748), anti-miRNA, and miR Synthetic were prepared and incubated at room temperature for 20 min (100 pM). Based on the cell groups, 1 µl of each miRNA/anti-miRNA solution and 1 µl of PEI solution were added into 200 µl of DEPC water and incubated at room temperature for 30 min. Then, the mixture was added to 1800 µl of additional culture medium on the sixth day43. After 4 h, the cells were washed with PBS buffer, re-cultured and, harvested after one day for RNA extraction.
A fluorescently labeled oligonucleotide (5′-6-FAM-CCCGAGACCCAACTGGTCACC -3′), as a green fluorescence emitter, was used to evaluate cellular delivery. Following the transfer of labeled oligonucleotide with PEI, the cells were incubated at 37 °C with 5% CO2 for 4 h before being washed with PBS buffer and imaged using an Olympus IX71 microscope (Fig. 1G).
Flow cytometry
The cell supernatant containing HSCs was centrifuged for 7 min at 1500 rpm. Then, the cells (3 × 104) were gently suspended in PBS buffer and treated using the labeled antibodies against CD90, CD45, and CD16 markers (Abcam in Cambridge, United Kingdom) for 25 min at room temperature (Fig. 1H).
RNA extraction, cDNA synthesis and RT-qPCR
After 24 h from the intervention period (4 h), the cells were counted and total RNA was extracted using the SinaPureTM ONE kit (Sinaclon, Tehran, Iran, Cat. No. EX6051). The Yektatajhiz cDNA Synthesis Kit (Tehran, Iran, Cat. No. YT4500) was used to synthesize the cDNA. The Real-time qRT-PCR technique was carried out using YTA SYBR Green qPCR Master mix (Yektatajhiz, Cat. No. YT2551). The GAPDH gene was used to normalize the gene expression levels. The reactions for all genes were run in 15 µl volumes. To design gene primers, the Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and OligoAnalyzer servers (https://eu.idtdna.com/pages/tools/oligoanalyzer) were used (Table 4).
Statistical analysis
The gene expression datasets were analyzed in R software (Version 4.3.0). The Kolmogorov–Smirnov test was used to assess the parametric and non-parametric distributions. Student's t-test, ANOVA, and Tukey’s post hoc test were used to compare the cellular groups in GraphPad Prism (Version 8.4.3). The flow cytometry data was analyzed with FlowJo software (Version 10). Using the 2−ΔΔCT formula, the changes in gene expression levels were estimated. P-values that were equal to or less than 0.05 were regarded as significant differences between the groups.
Ethics approval and consent to participate
The project titled “Systematic and functional studies of microRNA and anti-microRNA affecting genes involved in the growth process of hematopoietic stem cells isolated from human umbilical cord blood cells” was agreed by Research Ethics Committee of School of Medicine (IUMS). A research Ethics Committee certificate was presented and attached. (Ethics Co. IR.IUMS.FMD.REC.1402.048). The study was accomplished with a waiver for acquiring informed consent due to applying the unused human umbilical cord blood (UCB) bags.
Human and animal rights
Hematopoietic stem cells were prepared from the unused human umbilical cord blood (UCB) stored in the Iran Blood Transfusion Organization (IBTO).
Results
The prediction and experiment studies are shown in a flowchart (Fig. 1A–H).
RAS, PI3K-Akt, JAK-STAT, and cell cycle signaling pathways were predicted in the proliferation and differentiation processes of HSCs
The bioinformatics tools were used to analyze the gene data and to predict their roles in the cellular signaling pathways. The text mining analysis revealed 441 genes that were crucial for the differentiation and proliferation processes of HSCs. The major signaling pathways (KEGG) related to these genes included the PI3K-Akt signaling pathway (hsa04151, 73 genes), pathways in cancer (hsa05200, 66 genes), JAK-STAT signaling pathway (hsa04630, 62 genes), cell cycle (hsa04110, 59 genes), cytokine-cytokine receptor interaction (hsa04060, 59 genes), MAPK signaling pathway (hsa04010, 44 genes), Ras signaling pathway (hsa04014, 41 genes), Rap1 signaling pathway (hsa04015, 36 genes), and calcium signaling pathway (hsa04020, 27 genes). With annotating the suggested genes from the StemChecker database (Supplement 1 and 3), the four PI3K-Akt (2.36 E-109), RAS (1.85 E-42), JAK-STAT (1.43 E-73), and cell cycle (8.38 E-79) signaling pathways were more significant (Supplement 4) so that their genes were applied to generate the gene network in the STRING server (462 nodes, and 2221 edges) (Supplement 5A).
RB1, SMAD4, STAT1, CALML4, GNG13, and CDKN1A/CDKN1B genes, as well as miR-423-5p, miR-340-5p, miR-524-5p, and miR-20a-5p were predicted to control the major signaling pathways
The roles of miRNAs were predicted on the high score genes using the gene-miRNA network. The absolute values of fold data of HSC proliferation (n = 108) and differentiation (n = 103) genes estimated from GSE datasets (Supplement 2) were annotated to the nodes (green and pink colors) to improve and categorize the gene network. Some genes of the network that were not observed in the datasets according to the gene selection criteria were shown in light blue (Fig. 2). High- and low-fold genes including RB1 (+ 12.48), SMAD4 (+ 16.08), and STAT1 (+ 13.52) in the differentiation cluster (containing pink nodes) and CALML4 (+ 18.09), GNG13 (+ 17.97), CDKN1A (− 17.74), and CDKN1B (− 17.99) in the proliferation cluster (containing green nodes) were predicted on the gene network. Figure 3 displays the miRNAs related to the predicted genes. While miR-423-5p predicted to target both the CDKN1A and CDKN1B genes, miR-340-5p and miR-524-5p targeted the CALML4 and GNG13 genes. Furthermore, miR-20a-5p targeted the RB1, SMAD4, and STAT1 genes (Fig. 4).
Gene network. (A) The green nodes represents the genes involved in the proliferation process of HSCs. (B) The light blue nodes show the genes involved in the growth of HSCs but were not improved using the gene expression datasets. (C) The pink nodes represents the genes involved in the differentiation process. The node size is indicated as the absolute value of gene fold of expression datasets.
Heatmap graphs between miRNAs and target genes in the HSCs. (A) Top miRNAs targeting the CDKN1A and CDKN1B genes involved in the proliferation process. (B) Top miRNAs targeting the GNG13 and CALML4 genes related to the proliferation process. (C) Top miRNAs targeting the RB1, SMAD4, and STAT1 genes involved in the differentiation process.
The miRNA/anti-miRNA were efficiently transferred to HSCs by PEI
The cells were transfected with a FAM-ligated probe to evaluate the cell delivery. The fluorescent microscopic images showed that PEI transfers efficiently miRNA/anti-miRNA into the HSCs (Supplement 5B).
CD16, CD90, and CD45 were estimated during the study
The cells were analyzed with flow cytometry for the expression of CD90 and CD16 to determine the respective proportion of HSCs and differentiated cells on the first day after culturing the cells and on the seventh day. The CD16 and CD90 were estimated 17.9 ± 0.35% and 91.4 ± 0.65% one day after culturing the cells, respectively. However, their values changed conversely after the seventh day (CD16 31.1 ± 0.39%, CD90 17.7 ± 0.59%) (Supplement 5C) so that CD16 increased up to 1.73 times (p 0.035) while CD90 decreased by 5.16 times (p 0.0001). Also, the CD45 differentiation marker was estimated 78.6 ± 0.53% on the seventh day (Fig. 5A).
The miRNAs and anti-miRNA changed the cell count
The cell count was estimated as the proliferation rate of HSCs. The number of cells increased significantly in the PEI + (containing miR Synthetic, miR-20a-5p, miR-423-5p, and anti-miRNA) (2.66-fold, p 0.0001), PEI + miR-20a-5p (1.962-fold, p 0.0001), PEI + miR-423-5p (1.9-fold, p 0.0001) and PEI + anti-miRNA (1.43-fold, P 0.0001) groups as compared to the PEI + miR Synthetic group (Fig. 5B, Supplement 5D).
The miRNAs and anti-miRNA did not change CD45 marker
The changes of CD45 marker were compared between the cell groups as the differentiation rate of HSCs. There were no significant differences in the normalized CD45+/CD45- ratios among the groups. However, this ratio (0.877) was lower in the PEI + (miR-20a-5p, miR-423-5p, anti-miRNA, miR Synthetic) group. In other groups including PEI + miR-20a-5p, PEI + miR-423-5p, and PEI + anti-miRNA, the ratios were 0.912, 0.931, and 0.962, respectively (Fig. 5C, Supplement 5E).
The GNG13, CDKN1A, RB1, SMAD4, and STAT1 gene expression levels were changed using miRNAs and anti-miRNA
The gene expression levels were evaluated by the RT-qPCR technique. The CALML4 gene expression levels did not change significantly using anti-miRNA as compared to the PEI and PEI + miR Synthetic groups (p > 0.9) in the HSCs. However, the GNG13 gene expression levels increased significantly (p < 0.05) (Fig. 6A). The results also showed that CDKN1A gene expression levels decreased significantly in miR-423-5p/PEI-transfected cells as compared to the PEI + miR Synthetic and PEI groups (p 0.002 and p 0.001, respectively). However, the CDKN1B gene expression levels were not significantly changed in miR-423-5p/PEI-transfected cells (p > 0.8) (Fig. 6B). The RB1 gene expression levels decreased in the transfected cells with PEI + miR-20a-5p as compared to the transfected group with PEI + miR Synthetic (p 0.046). Similarly, the SMAD4 and STAT1 gene expression levels were lower in the miR-20a-5p/PEI-transfected cells (p 0.0001 and p 0.016, respectively) (Fig. 6C).
The gene expression changes in the HSCs. (A) The effects of anti-miRNA on the CALML4 and GNG13 genes. (B) The effects of miR-423-5p on the CDKN1A and CDKN1B gene expression levels. (C) The effects of miR-20a-5p on the RB1, SMAD4, and STAT1 gene expression levels. The data are presented as mean ± SD.
Discussion
HSCs are used in the treatment of blood disorders, metabolic deficiencies, and immune system abnormalities44. The standout features of umbilical cord blood HSCs include their non-invasive and painless blood collection methods, abundant presence of primitive and progenitor cells, capacity for cellular expansion, and high tolerance for the prevention of transplant rejection. Furthermore, HSCs can be used to regenerate damaged tissues, replace lost cells, and modulate the immune system. However, the limited number of HSCs may restrict the potential of these treatments45,46. It is well known that the expansion and differentiation of HSCs might be related to different internal and external factors. The external factors such as the microenvironment, adhesion molecules, growth factors, cytokines, chemical compounds, and co-culturing with other cells via the internal factors like signaling pathways can help to expand and differentiate HSCs4,13, 18, 47. Thus, the control of cell proliferation and differentiation processes of HSCs is an interesting subject48,49.
In this study, the data mining related to the proliferation and differentiation processes of HSCs were improved using the GEO datasets in agreement with the previous report50. In the proliferation stage, the GNG13 and CALML4 genes predicted the highest expression fold values, whereas CDKN1A and CDKN1B had the lowest. In the differentiation stage, the RB1, SMAD4, and STAT1 genes revealed the highest expression fold values. Based on the bioinformatics results, it was hypothesized to downregulate the CDKN1A, CDKN1B, STAT1, SMAD4, and RB1 genes and to upregulate the CALML4 and GNG13 genes for the improvement of proliferation and the prevention of differentiation of HSCs. Since some miRNAs were reported to regulate the genes involved in the proliferation and differentiation processes in HSCs51,52 thus in this study, the predicted genes were suggested to be targeted by miRNAs. The miR-20a-5p proposed the strongest ability to suppress the expression of three genes (RB1, SMAD4, and STAT1) in the differentiation process, while miR-423-5p suggested the highest potential to decrease the expression of two genes (CDKN1A and CDKN1B) that can inhibit proliferation stage. An anti-miRNA was designed to increase the expression of GNG13 and CALML4 genes by inhibiting miR-340-5p and miR-524-5p.
It was an effort to separate the non-adherent cells such as HSCs and hematopoietic progenitors through successive passages, separation, and culturing of supernatant cells at each stage41. Since attaching antibodies to cells and subjecting them to different reagents over an extended period can affect the typical states of the HSCs53 thus, it is crucial to minimize the loss of hematopoietic progenitor cells in a suitable microenvironment. For this reason, in line with the objective of this study to decrease the differentiation rate of HSCs and hematopoietic precursors while promoting their proliferation, a sequential partition purification method was employed and the cellular surface markers were evaluated. CD90 is a marker commonly expressed on many cells including HSCs, mesenchymal stem cells (MSCs), and keratinocytic stem cells (KSCs)54,55,56,57,58. The results suggested that the reduction of the CD90 marker (5 folds) following the sequential cell passages might be due to the omission of some cells including adherent stem cells on the seventh day. CD16 is also found in various differentiated cells such as natural killer cells, HSCs, neutrophils, monocytes, macrophages, and certain T cells59. Thus, its increase may occur because of the elevated differentiation of non-adherent cells such as HSCs on the seventh day. CD45 is present in various types of differentiated blood cells, excluding mature red blood cells and platelets60. Moreover, the predicted gene changes were evaluated on the seventh day since the CD16 and CD45 showed the cells shed their stemness conditions and developed into differentiated cells within seven days. The study results revealed that the anti-miRNA enhance the proliferation of HSCs. The anti-miRNA increased GNG13 gene expression levels, which may explain the reason for the enhanced cell proliferation. GNG13 is reported to be expressed in many tissues and contributes as a signal transmitter61,62. There were also many reports on the CDKN1A and CDKN1B genes and their impacts on cellular expansion. CDKN1A can inhibit cell cycle and tumor growth24. The increase in CDKN1A and CDKN1B gene expression levels maintained the quiescent state of HSCs and prevented the stem cell self-renewal63. Moreover, the growth of hepatocellular carcinoma is promoted by miR-423 via suppressing the expression of the tumor suppressor CDKN1A64. In agreement with these reports, the study results showed that the decrease in the CDKN1A gene expression by miR-423-5p improves the cell proliferation. In contrast with the gene-miRNA prediction results, however, there were no significant alterations due to the effects of anti-miRNA and miRNA on the CALML4 and CDKN1B gene expression levels. However, calcium is connected to CALML and is involved in the FGF, Ras, MAPK, and Wnt signaling pathways65. The activation of the MAPK pathway also affected the proliferation and differentiation of stem cells by a rise in the cytosolic calcium levels66. Some substances such as G-CSF, SCF, EPO, IL-3, and IL-6 increased the cellular calcium concentration leading to the proliferation and differentiation of the HSCs67.
The prediction results also suggested that the RB1, SMAD4, and STAT1 gene expression levels were elevated in the differentiation of HSCs. The experiment results showed the RB1, SMAD4, and STAT1 gene expression levels decreased by miR-20a-5p, in line with the gene-miRNA prediction results. Compared to the control group, however, the proliferation of HSCs was enhanced, but the CD45 marker, as a differentiation marker, had no change in the cell groups. Some studies agreed with our results on the involvement of RB1 in the cell cycle and differentiation68,69. Furthermore, the TGF-beta signaling pathway affected cellular growth via the SMAD4 isoforms attributing an antiproliferative response70. SMAD4 also regenerated and expanded the ability of human HSCs71. miR-130a decreased the sensitivity of TGF-β1-induced growth inhibition in a granulocytic cell line by suppressing the production of SMAD472. When differentiation-inducing cytokines bind to receptors on the surface of cells, they phosphorylate the STAT signaling molecule. The phosphorylated STATs (STAT1, STAT3, STAT5) combine to form dimers, which then move into the nucleus to regulate gene transcription. During all-trans retinoic acid (ATRA)-induced differentiation, STAT1 stops the cell cycle by regulating cyclins, p27, and c-Myc73,74. Megakaryopoiesis is significantly influenced by STAT175. On the other hand, miRNAs were able to change the gene expression levels. Some studies indicated a decrease in RB1 gene expression by miR-20a-5p76,77. The miR-20a-5p also suppressed the expression of SMAD4 in colorectal cancer cells78. The levels of miR-20a-5p may regulate the ability of human endometrium-derived mesenchymal stem cells (hEMSCs) to differentiate into cardiac cells, possibly by controlling the levels of SMAD478. The study results showed that in addition to miR-20a-5p, the combinational miRNA/anti-miRNA use elevates the proliferation process while preventing differentiation in HSCs.
Conclusion
The bioinformatics results predicted the list of genes and miRNAs related to the proliferation and differentiation stages of HSCs. Based on these data, some high-score genes and miRNAs were experimented in the HSCs. The study results confirmed that miR-423-5p and anti-miRNA promote the proliferation of HSCs by influencing the CDKN1A and GNG13 gene expression levels. Also, miR-20a-5p prevented the differentiation process in HSCs by suppressing the RB1, SMAD4, and STAT1 gene expression levels. Additionally, the combinational miR-423-5p/miR-20a-5p/anti-miRNA use caused a considerable increase in the proliferation process of HSCs.
Further studies are needed to understand the mechanism roles of miRNAs on the expression levels of suggested genes and to evaluate their protein values related to down- and upstream genes to gain knowledge of the signaling pathways involved in the proliferation and differentiation processes in the HSCs.
Data availability
The datasets analyzed during the current study are available in the GEO repository (https://www.ncbi.nlm.nih.gov/GEO). GSE107497, GSE125345, and GSE179928. All data are available in the main text or the supplementary materials.
References
Kolios, G. & Moodley, Y. Introduction to stem cells and regenerative medicine. Respiration. 85(1), 3–10 (2013).
Wisniewski, D., Affer, M., Willshire, J. & Clarkson, B. Further phenotypic characterization of the primitive lineage- CD34+CD38-CD90+CD45RA- hematopoietic stem cell/progenitor cell sub-population isolated from cord blood, mobilized peripheral blood and patients with chronic myelogenous leukemia. Blood Cancer J. 1(9), e36 (2011).
Cheng, H., Zheng, Z. & Cheng, T. New paradigms on hematopoietic stem cell differentiation. Protein Cell. 11(1), 34–44 (2020).
Walasek, M. A., van Os, R. & de Haan, G. Hematopoietic stem cell expansion: Challenges and opportunities. Ann. N. Y. Acad. Sci. 1266, 138–150 (2012).
Yu, B. et al. Co-expression of Runx1, Hoxa9, Hlf, and Hoxa7 confers multi-lineage potential on hematopoietic progenitors derived from pluripotent stem cells. Front Cell Dev. Biol. 10, 859769 (2022).
Lee, Y. et al. FGF signalling specifies haematopoietic stem cells through its regulation of somitic Notch signalling. Nat. Commun. 5, 5583 (2014).
Hofmeister, C. C., Zhang, J., Knight, K. L., Le, P. & Stiff, P. J. Ex vivo expansion of umbilical cord blood stem cells for transplantation: Growing knowledge from the hematopoietic niche. Bone Marrow Transplant. 39(1), 11–23 (2007).
Reya, T. et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 423(6938), 409–414 (2003).
Muzzey, D., Evans, E. A. & Lieber, C. Understanding the basics of NGS: From mechanism to variant calling. Curr. Genet. Med. Rep. 3(4), 158–165 (2015).
Wang, Y. & Blelloch, R. Cell cycle regulation by microRNAs in stem cells. Results Probl. Cell Differ. 53, 459–472 (2011).
Lima, J. F., Cerqueira, L., Figueiredo, C., Oliveira, C. & Azevedo, N. F. Anti-miRNA oligonucleotides: A comprehensive guide for design. RNA Biol. 15(3), 338–352 (2018).
Ajami, M., Soleimani, M., Abroun, S. & Atashi, A. Comparison of cord blood CD34 + stem cell expansion in coculture with mesenchymal stem cells overexpressing SDF-1 and soluble /membrane isoforms of SCF. J Cell Biochem. 120(9), 15297–15309 (2019).
Albayrak, E. & Kocabaş, F. Therapeutic targeting and HSC proliferation by small molecules and biologicals. Adv. Protein Chem. Struct. Biol. 135, 425–496 (2023).
Nakamura-Ishizu, A. Thrombopoietin regulates mitochondria homeostasis for hematopoietic stem cell maintenance. Rinsho Ketsueki. 62(5), 521–527. https://doi.org/10.11406/rinketsu.62.521 (2021) (Japanese).
Gao, P. et al. Transcriptional regulatory network controlling the ontogeny of hematopoietic stem cells. Genes Dev. 34(13–14), 950–964 (2020).
Kimura, T. & Yamazaki, S. Development of low-cost ex vivo hematopoietic stem cell expansion. Rinsho Ketsueki. 63(10), 1422–1429 (2022).
Gao, L., Decker, M., Chen, H. & Ding, L. Thrombopoietin from hepatocytes promotes hematopoietic stem cell regeneration after myeloablation. Elife. 31(10), e69894. https://doi.org/10.7554/eLife.69894.PMID:34463253;PMCID:PMC8457823 (2021).
Li, J. et al. Development and clinical advancement of small molecules for ex vivo expansion of hematopoietic stem cell. Acta Pharm Sin B. 12(6), 2808–2831 (2022).
Nakamura-Ishizu, A. & Suda, T. Multifaceted roles of thrombopoietin in hematopoietic stem cell regulation. Ann. N. Y. Acad. Sci. 1466(1), 51–58. https://doi.org/10.1111/nyas.14169 (2020).
Nakamura-Ishizu, A. et al. Prolonged maintenance of hematopoietic stem cells that escape from thrombopoietin deprivation. Blood. 137(19), 2609–2620 (2021).
Papa, L., Djedaini, M. & Hoffman, R. Ex vivo HSC expansion challenges the paradigm of unidirectional human hematopoiesis. Ann. N. Y. Acad. Sci. 1466(1), 39–50 (2020).
Lynch, J. et al. Hematopoietic stem cell quiescence and DNA replication dynamics maintained by the resilient β-catenin/Hoxa9/Prmt1 axis. Blood. 143(16), 1586–1598. https://doi.org/10.1182/blood.2023022082.PMID:38211335;PMCID:PMC11103100 (2024).
Sakamaki, T. et al. Hoxb5 defines the heterogeneity of self-renewal capacity in the hematopoietic stem cell compartment. Biochem. Biophys. Res. Commun. 539, 34–41 (2021).
Schirripa, A., Sexl, V. & Kollmann, K. Cyclin-dependent kinase inhibitors in malignant hematopoiesis. Front Oncol. 12, 916682 (2022).
Singh, A. K., Althoff, M. J. & Cancelas, J. A. Signaling pathways regulating hematopoietic stem cell and progenitor aging. Curr. Stem Cell Rep. 4(2), 166–181 (2018).
Wang, Y. & Sugimura, R. Ex vivo expansion of hematopoietic stem cells. Exp. Cell Res. 427(1), 113599 (2023).
Chen, Z., Guo, Q., Song, G. & Hou, Y. Molecular regulation of hematopoietic stem cell quiescence. Cell Mol. Life Sci. 79(4), 218. https://doi.org/10.1007/s00018-022-04200-w.PMID:35357574;PMCID:PMC11072845 (2022).
Zhang, L., Mack, R., Breslin, P. & Zhang, J. Molecular and cellular mechanisms of aging in hematopoietic stem cells and their niches. J. Hematol. Oncol. 13(1), 157 (2020).
Pinto, J. P. et al. StemChecker: A web-based tool to discover and explore stemness signatures in gene sets. Nucleic Acids Res. 43(W1), W72–W77 (2015).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51(D1), D587–D592 (2023).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11), 2498–2504 (2003).
Takagi, S. et al. Membrane-bound human SCF/KL promotes in vivo human hematopoietic engraftment and myeloid differentiation. Blood J. Am. Soc. Hematol. 119(12), 2768–2777 (2012).
Sigurjonsson, O. E., Gudmundsson, K. O., Haraldsdóttir, V., Rafnar, T. & Gudmundsson, S. Flt3/Flk-2-ligand in synergy with thrombopoietin delays megakaryocyte development and increases the numbers of megakaryocyte progenitor cells in serum-free cultures initiated with CD34+ cells. J. Hematotherapy Stem Cell Res. 11(2), 389–400 (2002).
Aizenman, Y. & de Vellis, J. Brain neurons develop in a serum and glial free environment: Effects of transferrin, insulin-insulin-like growth factor-I and thyroid hormone on neuronal survival, growth and differentiation. Brain Res. 406(1–2), 32–42 (1987).
Yadav, P., Vats, R., Bano, A. & Bhardwaj, R. Hematopoietic stem cells culture, expansion and differentiation: an insight into variable and available media. Int. J. Stem Cells. 13(3), 326–334 (2020).
Tsiftsoglou, A. S. Erythropoietin (EPO) as a key regulator of erythropoiesis, bone remodeling and endothelial transdifferentiation of multipotent mesenchymal stem cells (MSCs): Implications in regenerative medicine. Cells. 10(8), 2140 (2021).
Nogueira-Pedro, A. et al. α-Tocopherol induces hematopoietic stem/progenitor cell expansion and ERK1/2-mediated differentiation. J. Leukocyte Biol. 90(6), 1111–1117 (2011).
Hu, Q. et al. GPX4 and vitamin E cooperatively protect hematopoietic stem and progenitor cells from lipid peroxidation and ferroptosis. Cell Death Dis. 12(7), 706 (2021).
Ren, Y., Cui, Y. N. & Wang, H. W. Effects of different concentrations of nicotinamide on hematopoietic stem cells cultured in vitro. World J. Stem Cells. 16(2), 163–175 (2024).
Brewer, G. J., Torricelli, J. R., Evege, E. K. & Price, P. J. Optimized survival of hippocampal neurons in B27-supplemented neurobasal™, a new serum-free medium combination. J. Neurosci. Res. 35(5), 567–576 (1993).
Phuc, P. V. et al. Isolation of three important types of stem cells from the same samples of banked umbilical cord blood. Cell Tissue Bank. 13(2), 341–351 (2012).
Grassinger, J. et al. Differentiation of hematopoietic progenitor cells towards the myeloid and B-lymphoid lineage by hepatocyte growth factor (HGF) and thrombopoietin (TPO) together with early acting cytokines. Eur. J. Haematol. 77(2), 134–144 (2006).
Rayner, K. J. & Moore, K. J. The plaque “micro” environment: microRNAs control the risk and the development of atherosclerosis. Curr. Atheroscler. Rep. 14(5), 413–421 (2012).
Alexander, T., Greco, R. & Snowden, J. A. Hematopoietic stem cell transplantation for autoimmune disease. Annu. Rev. Med. 72, 215–228 (2021).
Zhu, X., Tang, B. & Sun, Z. Umbilical cord blood transplantation: Still growing and improving. Stem Cells Transl. Med. 10(Suppl 2), S62-s74 (2021).
Gudauskaitė, G., Kairienė, I., Ivaškienė, T., Rascon, J. & Mobasheri, A. Therapeutic perspectives for the clinical application of umbilical cord hematopoietic and mesenchymal stem cells: Overcoming complications arising after allogeneic hematopoietic stem cell transplantation. Adv. Exp. Med. Biol. 1409, 111–126 (2023).
Wilkinson, A. C., Igarashi, K. J. & Nakauchi, H. Haematopoietic stem cell self-renewal in vivo and ex vivo. Nat. Rev. Genet. 21(9), 541–554 (2020).
Amiri, F., Kiani, A. A., Bahadori, M. & Roudkenar, M. H. Co-culture of mesenchymal stem cell spheres with hematopoietic stem cells under hypoxia: A cost-effective method to maintain self-renewal and homing marker expression. Mol. Biol. Rep. 49(2), 931–941 (2022).
Shirdare, M., Amiri, F., Samiee, M. P. & Safari, A. Influential factors for optimizing and strengthening mesenchymal stem cells and hematopoietic stem cells co-culture. Mol. Biol. Rep. 51(1), 189 (2024).
Elahimanesh, M., Shokri, N., Mohammadi, P., Parvaz, N. & Najafi, M. Step by step analysis on gene datasets of growth phases in hematopoietic stem cells. Biochem. Biophys. Rep. 1(39), 101737 (2024).
Diener, C., Keller, A. & Meese, E. Emerging concepts of miRNA therapeutics: from cells to clinic. Trends Genet. 38(6), 613–626 (2022).
Nowicki, M. et al. Alterations in microRNA expression during hematopoietic stem cell mobilization. Biology (Basel). 10(7), 668 (2021).
Hu, P., Zhang, W., Xin, H. & Deng, G. Single cell isolation and analysis. Front Cell Dev. Biol. 4, 116 (2016).
Papa, L. et al. Ex vivo human HSC expansion requires coordination of cellular reprogramming with mitochondrial remodeling and p53 activation. Blood Adv. 2(20), 2766–2779 (2018).
Hua, P. et al. Single-cell assessment of transcriptome alterations induced by Scriptaid in early differentiated human haematopoietic progenitors during ex vivo expansion. Sci. Rep. 9(1), 5300 (2019).
Radtke, S. et al. Purification of human CD34(+)CD90(+) HSCs reduces target cell population and improves lentiviral transduction for gene therapy. Mol. Ther. Methods Clin. Dev. 18, 679–691 (2020).
Rix, B., Maduro, A. H., Bridge, K. S. & Grey, W. Markers for human haematopoietic stem cells: The disconnect between an identification marker and its function. Front Physiol. 13, 1009160 (2022).
Fromm, P. D. et al. Distinguishing human peripheral blood CD16(+) myeloid cells based on phenotypic characteristics. J. Leukoc. Biol. 107(2), 323–339 (2020).
Rheinländer, A., Schraven, B. & Bommhardt, U. CD45 in human physiology and clinical medicine. Immunol. Lett. 196, 22–32 (2018).
Sanfilippo, C. et al. GNG13 is a potential marker of the state of health of Alzheimer’s disease patients’ cerebellum. J. Mol. Neurosci. 71, 1046–1060 (2021).
Liu, Z. et al. Identification of new binding partners of the chemosensory signaling protein Gγ13 expressed in taste and olfactory sensory cells. Front. Cell. Neurosci. 6, 26 (2012).
Jia, W. et al. Indispensable role of Galectin-3 in promoting quiescence of hematopoietic stem cells. Nat Commun. 12(1), 2118 (2021).
Lin, J. et al. MicroRNA-423 promotes cell growth and regulates G(1)/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis. 32(11), 1641–1647 (2011).
Tonelli, F. M. P. et al. Stem cells and calcium signaling. In Calcium signaling (ed. Islam, M. S.) 891–916 (Springer, 2012).
Santos, E. W. et al. Protein restriction impairs the response activation/responsivity of MAPK signaling pathway of hematopoietic stem cells. Nutr. Res. 116, 12–23 (2023).
Uslu, M., Albayrak, E. & Kocabaş, F. Temporal modulation of calcium sensing in hematopoietic stem cells is crucial for proper stem cell expansion and engraftment. J. Cell Physiol. 235(12), 9644–9666 (2020).
Furukawa, Y. et al. Expression and state of phosphorylation of the retinoblastoma susceptibility gene product in cycling and noncycling human hematopoietic cells. Proc. Natl. Acad. Sci. U. S. A. 87(7), 2770–2774 (1990).
Mushtaq, M., Gaza, H. V. & Kashuba, E. V. Role of the RB-interacting proteins in stem cell biology. Adv. Cancer Res. 131, 133–157 (2016).
He, W. et al. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 125(5), 929–941 (2006).
Rörby, E., Hägerström, M. N., Blank, U., Karlsson, G. & Karlsson, S. Human hematopoietic stem/progenitor cells overexpressing Smad4 exhibit impaired reconstitution potential in vivo. Blood. 120(22), 4343–4351 (2012).
Häger, M. et al. MicroRNA-130a-mediated down-regulation of Smad4 contributes to reduced sensitivity to TGF-β1 stimulation in granulocytic precursors. Blood. 118(25), 6649–6659 (2011).
Miranda, M. B. & Johnson, D. E. Signal transduction pathways that contribute to myeloid differentiation. Leukemia. 21(7), 1363–1377 (2007).
Dimberg, A., Karlberg, I., Nilsson, K. & Oberg, F. Ser727/Tyr701-phosphorylated Stat1 is required for the regulation of c-Myc, cyclins, and p27Kip1 associated with ATRA-induced G0/G1 arrest of U-937 cells. Blood. 102(1), 254–261 (2003).
Fasouli, E. S. & Katsantoni, E. JAK-STAT in early hematopoiesis and leukemia. Front Cell Dev. Biol. 9, 669363 (2021).
Liu, H. et al. The SOX4/miR-17-92/RB1 axis promotes prostate cancer progression. Neoplasia. 21(8), 765–776 (2019).
Chen, Y. J., Hsu, C. T., Tsai, S. F. & Chen, C. H. Association between circulating MicroRNAs (miR-21–5p, miR-20a-5p, miR-29b-3p, miR-126–3p and miR-101–3p) and chronic allograft dysfunction in renal transplant recipients. Int. J. Mol. Sci. 23(20), 12253 (2022).
Cheng, D. et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget. 7(29), 45199–45213 (2016).
Maleki, B. et al. Effect of miR-18a-5p, miR-19a-3p, and miR-20a-5p on in vitro cardiomyocyte differentiation of human endometrium tissue-derived stem cells through regulation of Smad4 expression. Rep. Biochem. Mol. Biol. 12(1), 136–146 (2023).
Acknowledgements
The authors thank the staff of the Iran Blood Transfusion Organization (IBTO).
Funding
It is supported by Iran University of Medical Sciences (No. 1401-4-4-24931).
Author information
Authors and Affiliations
Contributions
MN and ME design of study. NS, MR, RS and ME collected and analyzed. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Elahimanesh, M., Shokri, N., Shabani, R. et al. Exploring the potential of predicted miRNAs on the genes involved in the expansion of hematopoietic stem cells. Sci Rep 14, 15551 (2024). https://doi.org/10.1038/s41598-024-66614-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-66614-9
- Springer Nature Limited