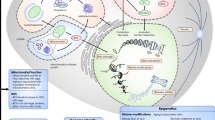
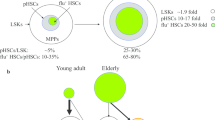
Abstract
Purpose of Review
The functional decline of hematopoiesis that occurs in the elderly, or in patients who receive therapies that trigger cellular senescence effects, results in a progressive reduction in the immune response and an increased incidence of myeloid malignancy. Intracellular signals in hematopoietic stem cells and progenitors (HSC/P) mediate systemic, microenvironment, and cell-intrinsic effector aging signals that induce their decline. This review intends to summarize and critically review our advances in the understanding of the intracellular signaling pathways responsible for HSC decline during aging and opportunities for intervention.
Recent Findings
For a long time, aging of HSC has been thought to be an irreversible process imprinted in stem cells due to the cell-intrinsic nature of aging. However, recent murine models and human correlative studies provide evidence that aging is associated with molecular signaling pathways, including oxidative stress, metabolic dysfunction, loss of polarity, and an altered epigenome. These signaling pathways provide potential targets for prevention or reversal of age-related changes.
Summary
Here, we review our current understanding of the signaling pathways that are differentially activated or repressed during HSC/P aging, focusing on the oxidative, metabolic, biochemical, and structural consequences downstream, and cell-intrinsic, systemic, and environmental influences.


Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Cheng J, Turkel N, Hemati N, Fuller MT, Hunt AJ, Yamashita YM. Centrosome misorientation reduces stem cell division during aging. Nature. 2008;456(7222):599–604. https://doi.org/10.1038/nature07386.
• Bernitz JM, Kim HS, MacArthur B, Sieburg H, Moore K. Hematopoietic stem cells count and remember self-renewal divisions. Cell. 2016;167(5):1296–309 e10. https://doi.org/10.1016/j.cell.2016.10.022. This article provides evidence that demonstrates age-related phenotypic changes within the HSC compartment are divisional history dependent.
Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135(6):1118–29. https://doi.org/10.1016/j.cell.2008.10.048.
Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441(7097):1068–74. https://doi.org/10.1038/nature04956.
Bryder D, Rossi DJ, Weissman IL. Hematopoietic stem cells: the paradigmatic tissue-specific stem cell. Am J Pathol. 2006;169(2):338–46. https://doi.org/10.2353/ajpath.2006.060312.
Liang Y, Van Zant G, Szilvassy SJ. Effects of aging on the homing and engraftment of murine hematopoietic stem and progenitor cells. Blood. 2005;106(4):1479–87. https://doi.org/10.1182/blood-2004-11-4282.
Morrison SJ, Wandycz AM, Akashi K, Globerson A, Weissman IL. The aging of hematopoietic stem cells. Nat Med. 1996;2(9):1011–6.
Rossi DJ, Bryder D, Zahn JM, Ahlenius H, Sonu R, Wagers AJ, et al. Cell intrinsic alterations underlie hematopoietic stem cell aging. Proc Natl Acad Sci U S A. 2005;102(26):9194–9. https://doi.org/10.1073/pnas.0503280102.
Xing Z, Ryan MA, Daria D, Nattamai KJ, Van Zant G, Wang L, et al. Increased hematopoietic stem cell mobilization in aged mice. Blood. 2006;108(7):2190–7. https://doi.org/10.1182/blood-2005-12-010272.
Geiger H, Rudolph KL. Aging in the lympho-hematopoietic stem cell compartment. Trends Immunol. 2009;30(7):360–5. https://doi.org/10.1016/j.it.2009.03.010.
de Haan G, Nijhof W, Van Zant G. Mouse strain-dependent changes in frequency and proliferation of hematopoietic stem cells during aging: correlation between lifespan and cycling activity. Blood. 1997;89(5):1543–50.
de Haan G, Van Zant G. Dynamic changes in mouse hematopoietic stem cell numbers during aging. Blood. 1999;93(10):3294–301.
Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127(2):265–75. https://doi.org/10.1016/j.cell.2006.10.003.
Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: the critical roles of p53. Oncogene. 2013;32(43):5129–43. https://doi.org/10.1038/onc.2012.640.
Geiger H, de Haan G, Florian MC. The aging haematopoietic stem cell compartment. Nat Rev Immunol. 2013;13(5):376–89. https://doi.org/10.1038/nri3433.
Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med. 2014;20(8):833–46. https://doi.org/10.1038/nm.3647.
Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, et al. Stem-cell aging modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443(7110):421–6. https://doi.org/10.1038/nature05159.
Jeck WR, Siebold AP, Sharpless NE. Review: a meta-analysis of GWAS and age-associated diseases. Aging Cell. 2012;11(5):727–31. https://doi.org/10.1111/j.1474-9726.2012.00871.x.
• Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–9. https://doi.org/10.1038/nature16932. This paper provides evidence of p16Ink4a positive cells accumulation with aging and it removal as an important therapeutic approach to extend longevity
Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–5. https://doi.org/10.1038/nature01587.
Attema JL, Pronk CJ, Norddahl GL, Nygren JM, Bryder D. Hematopoietic stem cell aging is uncoupled from p16 INK4A-mediated senescence. Oncogene. 2009;28(22):2238–43. https://doi.org/10.1038/onc.2009.94.
• Chang J, Wang Y, Shao L, Laberge RM, Demaria M, Campisi J, et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat Med. 2016;22(1):78–83. https://doi.org/10.1038/nm.4010. This article shows that selective clearance of senescent cells rejuvenate aged stem cells.
•• Zhang H, Ryu D. Wu Y, Gariani K, Wang X, Luan P et al. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science. 2016;352(6292):1436–43. https://doi.org/10.1126/science.aaf2693. This article demonstrates that NAD+ supplemantation delays senescence and rejuvanate aged HSC through induction of mitochondrial unfolded protein response and synthesis of prohibitin proteins.
Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32(1):2–11. https://doi.org/10.1128/MCB.06159-11.
Kennedy BK, Lamming DW. The mechanistic target of rapamycin: the grand conducTOR of metabolism and aging. Cell Metab. 2016;23(6):990–1003. https://doi.org/10.1016/j.cmet.2016.05.009.
Florian MC, Dorr K, Niebel A, Daria D, Schrezenmeier H, Rojewski M, et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell. 2012;10(5):520–30. https://doi.org/10.1016/j.stem.2012.04.007.
Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12(4):446–51. https://doi.org/10.1038/nm1388.
Jang YY, Sharkis SJ. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 2007;110(8):3056–63. https://doi.org/10.1182/blood-2007-05-087759.
Lopez-Otin C, Galluzzi L, Freije JMP, Madeo F, Kroemer G. Metabolic control of longevity. Cell. 2016;166(4):802–21. https://doi.org/10.1016/j.cell.2016.07.031.
Cheng CW, Adams GB, Perin L, Wei M, Zhou X, Lam BS, et al. Prolonged fasting reduces IGF-1/PKA to promote hematopoietic-stem-cell-based regeneration and reverse immunosuppression. Cell Stem Cell. 2014;14(6):810–23. https://doi.org/10.1016/j.stem.2014.04.014.
Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of aging and age-related disease. Nature. 2013;493(7432):338–45. https://doi.org/10.1038/nature11861.
Satoh Y, Yokota T, Sudo T, Kondo M, Lai A, Kincade PW, et al. The Satb1 protein directs hematopoietic stem cell differentiation toward lymphoid lineages. Immunity. 2013;38(6):1105–15. https://doi.org/10.1016/j.immuni.2013.05.014.
Brown K, Xie S, Qiu X, Mohrin M, Shin J, Liu Y, et al. SIRT3 reverses aging-associated degeneration. Cell Rep. 2013;3(2):319–27. https://doi.org/10.1016/j.celrep.2013.01.005.
•• Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, et al. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 2015;347(6228):1374–7. https://doi.org/10.1126/science.aaa2361. This study identified SIRT7-NRF1 as a regulator of mitochondrial unfolded protein response and define its significance in rejuvanation of aged HSC.
Hall BM, Balan V, Gleiberman AS, Strom E, Krasnov P, Virtuoso LP, et al. p16(Ink4a) and senescence-associated beta-galactosidase can be induced in macrophages as part of a reversible response to physiological stimuli. Aging (Albany NY). 2017;9(8):1867–84. https://doi.org/10.18632/aging.101268.
Hall BM, Balan V, Gleiberman AS, Strom E, Krasnov P, Virtuoso LP, et al. Aging of mice is associated with p16(Ink4a)- and beta-galactosidase-positive macrophage accumulation that can be induced in young mice by senescent cells. Aging (Albany NY). 2016;8(7):1294–315. https://doi.org/10.18632/aging.100991.
Carrasco-Garcia E, Moreno M, Moreno-Cugnon L, Matheu A. Increased Arf/p53 activity in stem cells, aging and cancer. Aging Cell. 2017;16(2):219–25. https://doi.org/10.1111/acel.12574.
Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, et al. p53 mutant mice that display early aging-associated phenotypes. Nature. 2002;415(6867):45–53. https://doi.org/10.1038/415045a.
Maier B, Gluba W, Bernier B, Turner T, Mohammad K, Guise T, et al. Modulation of mammalian life span by the short isoform of p53. Genes Dev. 2004;18(3):306–19. https://doi.org/10.1101/gad.1162404.
Dumble M, Moore L, Chambers SM, Geiger H, Van Zant G, Goodell MA, et al. The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood. 2007;109(4):1736–42. https://doi.org/10.1182/blood-2006-03-010413.
Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, et al. Delayed aging through damage protection by the Arf/p53 pathway. Nature. 2007;448(7151):375–9. https://doi.org/10.1038/nature05949.
Armata HL, Garlick DS, Sluss HK. The ataxia telangiectasia-mutated target site Ser18 is required for p53-mediated tumor suppression. Cancer Res. 2007;67(24):11696–703. https://doi.org/10.1158/0008-5472.CAN-07-1610.
Liu D, Ou L, Clemenson GD Jr, Chao C, Lutske ME, Zambetti GP, et al. Puma is required for p53-induced depletion of adult stem cells. Nat Cell Biol. 2010;12(10):993–8. https://doi.org/10.1038/ncb2100.
Domen J, Cheshier SH, Weissman IL. The role of apoptosis in the regulation of hematopoietic stem cells: Overexpression of Bcl-2 increases both their number and repopulation potential. J Exp Med. 2000;191(2):253–64.
•• Kirschner K, Chandra T, Kiselev V, Flores-Santa Cruz D, Macaulay IC, Park HJ, et al. Proliferation drives aging-related functional decline in a subpopulation of the hematopoietic stem cell compartment. Cell Rep. 2017;19(8):1503–11. https://doi.org/10.1016/j.celrep.2017.04.074. This article defines cellular heterogeneity in HSC aging, and highlighted the significance of JAK/STAT signaling in stem cell exhaustion.
Kim HN, Chang J, Shao L, Han L, Iyer S, Manolagas SC, et al. DNA damage and senescence in osteoprogenitors expressing Osx1 may cause their decrease with age. Aging Cell. 2017;16(4):693–703. https://doi.org/10.1111/acel.12597.
Beerman I, Bhattacharya D, Zandi S, Sigvardsson M, Weissman IL, Bryder D, et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci U S A. 2010;107(12):5465–70. https://doi.org/10.1073/pnas.1000834107.
Dykstra B, Olthof S, Schreuder J, Ritsema M, de Haan G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J Exp Med. 2011;208(13):2691–703. https://doi.org/10.1084/jem.20111490.
Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518(7540):542–6. https://doi.org/10.1038/nature14242.
Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho YJ, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514(7522):322–7. https://doi.org/10.1038/nature13824.
Verovskaya E, Broekhuis MJ, Zwart E, Ritsema M, van Os R, de Haan G, et al. Heterogeneity of young and aged murine hematopoietic stem cells revealed by quantitative clonal analysis using cellular barcoding. Blood. 2013;122(4):523–32. https://doi.org/10.1182/blood-2013-01-481135.
•• Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–87. https://doi.org/10.1056/NEJMoa1409405. This article preformed whole exome sequencing on a large cohort and reveiled detectable clonal expansions most frequently involved somatic mutations in three genes (DNMT3A, ASXL1, and TET2) that have previously been implicated in hematologic cancers
•• Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–98. https://doi.org/10.1056/NEJMoa1408617. This article highlights that detectable somatic mutations, partcularly in DNMT3A, TET2, and ASXL1, were rare in persons younger than 40 years of age but rose appreciably in frequency with age and was associated with an increase in the risk of hematologic cancer, an increase in all-cause mortality, with increases in the risks of incident coronary heart disease and ischemic stroke.
•• McKerrell T, Park N, Moreno T, Grove CS, Ponstingl H, Stephens J, et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10(8):1239–45. https://doi.org/10.1016/j.celrep.2015.02.005. This paper indicates that spliceosome gene mutations drive clonal expansion under selection pressures particular to the aging hemopoietic system and explains the high incidence of clonal disorders associated with these mutations in advanced old age.
•• Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. https://doi.org/10.1182/blood-2015-03-631747. This article highlights the acquisition of somatic mutations that drive clonal expansion in the absence of cytopenias and dysplastic hematopoiesis can be considered clonal hematopoiesis of indeterminate potential (CHIP).
Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 2014;15(1):37–50. https://doi.org/10.1016/j.stem.2014.04.016.
Rossi DJ, Seita J, Czechowicz A, Bhattacharya D, Bryder D, Weissman IL. Hematopoietic stem cell quiescence attenuates DNA damage response and permits DNA damage accumulation during aging. Cell Cycle. 2007;6(19):2371–6. https://doi.org/10.4161/cc.6.19.4759.
Flach J, Bakker ST, Mohrin M, Conroy PC, Pietras EM, Reynaud D, et al. Replication stress is a potent driver of functional decline in aging haematopoietic stem cells. Nature. 2014;512(7513):198–202. https://doi.org/10.1038/nature13619.
•• Walter D, Lier A, Geiselhart A, Thalheimer FB, Huntscha S, Sobotta MC, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520(7548):549–52. https://doi.org/10.1038/nature14131. This article establishes a novel link between physiological stress and DNA damage in normal HSC while providing a mechanistic explanation for the accumulation of DNA damage in HSC during aging and the accelerated failure of the haematopoietic system in Fanconi anaemia patients.
Seita J, Rossi DJ, Weissman IL. Differential DNA damage response in stem and progenitor cells. Cell Stem Cell. 2010;7(2):145–7. https://doi.org/10.1016/j.stem.2010.07.006.
Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, et al. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21(19):2495–508. https://doi.org/10.1101/gad.439107.
Wahlestedt M, Norddahl GL, Sten G, Ugale A, Frisk MA, Mattsson R, et al. An epigenetic component of hematopoietic stem cell aging amenable to reprogramming into a young state. Blood. 2013;121(21):4257–64. https://doi.org/10.1182/blood-2012-11-469080.
Morrison SJ, Prowse KR, Ho P, Weissman IL. Telomerase activity in hematopoietic cells is associated with self-renewal potential. Immunity. 1996;5(3):207–16.
Allsopp RC, Cheshier S, Weissman IL. Telomere shortening accompanies increased cell cycle activity during serial transplantation of hematopoietic stem cells. J Exp Med. 2001;193(8):917–24.
Allsopp RC, Morin GB, Horner JW, DePinho R, Harley CB, Weissman IL. Effect of TERT over-expression on the long-term transplantation capacity of hematopoietic stem cells. Nat Med. 2003;9(4):369–71. https://doi.org/10.1038/nm0403-369.
Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5(8):e201. https://doi.org/10.1371/journal.pbio.0050201.
Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, et al. DNA repair is limiting for haematopoietic stem cells during aging. Nature. 2007;447(7145):686–90. https://doi.org/10.1038/nature05875.
Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447(7145):725–9. https://doi.org/10.1038/nature05862.
Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–8. https://doi.org/10.1038/nature08467.
Kenyon J, Gerson SL. The role of DNA damage repair in aging of adult stem cells. Nucleic Acids Res. 2007;35(22):7557–65. https://doi.org/10.1093/nar/gkm1064.
Qing Y, Wang Z, Bunting KD, Gerson SL. Bcl2 overexpression rescues the hematopoietic stem cell defects in Ku70-deficient mice by restoration of quiescence. Blood. 2014;123(7):1002–11. https://doi.org/10.1182/blood-2013-08-521716.
Prall WC, Czibere A, Jager M, Spentzos D, Libermann TA, Gattermann N, et al. Age-related transcription levels of KU70, MGST1 and BIK in CD34+ hematopoietic stem and progenitor cells. Mech Aging Dev. 2007;128(9):503–10. https://doi.org/10.1016/j.mad.2007.06.008.
Bender CF, Sikes ML, Sullivan R, Huye LE, Le Beau MM, Roth DB, et al. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev. 2002;16(17):2237–51. https://doi.org/10.1101/gad.1007902.
Prasher JM, Lalai AS, Heijmans-Antonissen C, Ploemacher RE, Hoeijmakers JH, Touw IP, et al. Reduced hematopoietic reserves in DNA interstrand crosslink repair-deficient Ercc1−/− mice. EMBO J. 2005;24(4):861–71. https://doi.org/10.1038/sj.emboj.7600542.
Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86(1):159–71.
Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431(7011):997–1002. https://doi.org/10.1038/nature02989.
Moehrle BM, Nattamai K, Brown A, Florian MC, Ryan M, Vogel M, et al. Stem cell-specific mechanisms ensure genomic fidelity within HSCs and upon aging of HSCs. Cell Rep. 2015;13(11):2412–24. https://doi.org/10.1016/j.celrep.2015.11.030.
Takubo K, Nagamatsu G, Kobayashi CI, Nakamura-Ishizu A, Kobayashi H, Ikeda E, et al. Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell. 2013;12(1):49–61. https://doi.org/10.1016/j.stem.2012.10.011.
Hamilton ML, Van Remmen H, Drake JA, Yang H, Guo ZM, Kewitt K, et al. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci U S A. 2001;98(18):10469–74. https://doi.org/10.1073/pnas.171202698.
Kolosova NG, Stefanova NA, Muraleva NA, Skulachev VP. The mitochondria-targeted antioxidant SkQ1 but not N-acetylcysteine reverses aging-related biomarkers in rats. Aging (Albany NY). 2012;4(10):686–94. https://doi.org/10.18632/aging.100493.
Juntilla MM, Patil VD, Calamito M, Joshi RP, Birnbaum MJ, Koretzky GA. AKT1 and AKT2 maintain hematopoietic stem cell function by regulating reactive oxygen species. Blood. 2010;115(20):4030–8. https://doi.org/10.1182/blood-2009-09-241000.
Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Liu Y, et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205(10):2397–408. https://doi.org/10.1084/jem.20081297.
Borodkina A, Shatrova A, Abushik P, Nikolsky N, Burova E. Interaction between ROS dependent DNA damage, mitochondria and p38 MAPK underlies senescence of human adult stem cells. Aging (Albany NY). 2014;6(6):481–95. https://doi.org/10.18632/aging.100673.
Jung H, Kim DO, Byun JE, Kim WS, Kim MJ, Song HY, et al. Thioredoxin-interacting protein regulates haematopoietic stem cell aging and rejuvenation by inhibiting p38 kinase activity. Nat Commun. 2016;7:13674. https://doi.org/10.1038/ncomms13674.
Wang Y, Hekimi S. Mitochondrial dysfunction and longevity in animals: Untangling the knot. Science. 2015;350(6265):1204–7. https://doi.org/10.1126/science.aac4357.
Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. https://doi.org/10.1016/j.cell.2013.05.039.
Norddahl GL, Pronk CJ, Wahlestedt M, Sten G, Nygren JM, Ugale A, et al. Accumulating mitochondrial DNA mutations drive premature hematopoietic aging phenotypes distinct from physiological stem cell aging. Cell Stem Cell. 2011;8(5):499–510. https://doi.org/10.1016/j.stem.2011.03.009.
Liang R, Ghaffari S. Mitochondria and FOXO3 in stem cell homeostasis, a window into hematopoietic stem cell fate determination. J Bioenerg Biomembr. 2017;49(4):343–6. https://doi.org/10.1007/s10863-017-9719-7.
Kohli L, Passegue E. Surviving change: the metabolic journey of hematopoietic stem cells. Trends Cell Biol. 2014;24(8):479–87. https://doi.org/10.1016/j.tcb.2014.04.001.
Luchsinger LL, de Almeida MJ, Corrigan DJ, Mumau M, Snoeck HW. Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature. 2016;529(7587):528–31. https://doi.org/10.1038/nature16500.
Sun N, Youle RJ, Finkel T. The mitochondrial basis of aging. Mol Cell. 2016;61(5):654–66. https://doi.org/10.1016/j.molcel.2016.01.028.
Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, et al. Premature aging in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–23. https://doi.org/10.1038/nature02517.
Oh J, Lee YD, Wagers AJ. Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nature Medicine. 2014;20(8):870–80. https://doi.org/10.1038/nm.3651.
Fang EF, Lautrup S, Hou Y, Demarest TG, Croteau DL, Mattson MP, et al. NAD+ in aging: molecular mechanisms and translational implications. Trends in Mol Med. 2017;23(10):899–916. https://doi.org/10.1016/j.molmed.2017.08.001.
Bernales S, Soto MM, McCullagh E. Unfolded protein stress in the endoplasmic reticulum and mitochondria: a role in neurodegeneration. Front Aging Neurosci. 2012;4:5. https://doi.org/10.3389/fnagi.2012.00005.
Shyh-Chang N, Daley GQ, Cantley LC. Stem cell metabolism in tissue development and aging. Development. 2013;140(12):2535–47. https://doi.org/10.1242/dev.091777.
Schreiber KH, Kennedy BK. When lamins go bad: nuclear structure and disease. Cell. 2013;152(6):1365–75. https://doi.org/10.1016/j.cell.2013.02.015.
Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326(5949):140–4. https://doi.org/10.1126/science.1177221.
Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, et al. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 2013;4(5):913–20. https://doi.org/10.1016/j.celrep.2013.07.030.
Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335(6076):1638–43. https://doi.org/10.1126/science.1215135.
Rimmele P, Bigarella CL, Liang R, Izac B, Dieguez-Gonzalez R, Barbet G, et al. Aging-like phenotype and defective lineage specification in SIRT1-deleted hematopoietic stem and progenitor cells. Stem Cell Reports. 2014;3(1):44–59. https://doi.org/10.1016/j.stemcr.2014.04.015.
O'Callaghan C, Vassilopoulos A. Sirtuins at the crossroads of stemness, aging, and cancer. Aging Cell. 2017;16(6):1208–18. https://doi.org/10.1111/acel.12685.
•• Wang H, Diao D, Shi Z, Zhu X, Gao Y, Gao S, et al. SIRT6 controls hematopoietic stem cell homeostasis through epigenetic regulation of Wnt signaling. Cell Stem Cell. 2016;18(4):495–507. https://doi.org/10.1016/j.stem.2016.03.005. This study for the first time shows the link between SIRT6 and Wnt signaling in the regulation of HSC homeostasis and self-renewal.
Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17(1):41–52. https://doi.org/10.1016/j.ccr.2009.11.023.
Wrighton KH. Stem cells:SIRT7, the UPR and HSC aging. Nat Rev Mol Cell Biol. 2015;16(5):266–7. https://doi.org/10.1038/nrm3981.
Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155(7):1624–38. https://doi.org/10.1016/j.cell.2013.11.037.
Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–29. https://doi.org/10.1016/j.cell.2005.11.044.
Tothova Z, Gilliland DG. FoxO transcription factors and stem cell homeostasis: insights from the hematopoietic system. Cell Stem Cell. 2007;1(2):140–52. https://doi.org/10.1016/j.stem.2007.07.017.
Mohrin M, Chen D. The mitochondrial metabolic checkpoint and aging of hematopoietic stem cells. Curr Opin Hematol. 2016;23(4):318–24. https://doi.org/10.1097/MOH.0000000000000244.
Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15(4):243–56. https://doi.org/10.1038/nrm3772.
Bigarella CL, Li J, Rimmele P, Liang R, Sobol RW, Ghaffari S. FOXO3 transcription factor is essential for protecting hematopoietic stem and progenitor cells from oxidative DNA damage. J Biol Chem. 2017;292(7):3005–15. https://doi.org/10.1074/jbc.M116.769455.
Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128(2):325–39. https://doi.org/10.1016/j.cell.2007.01.003.
Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1(1):101–12. https://doi.org/10.1016/j.stem.2007.02.001.
Rimmele P, Liang R, Bigarella CL, Kocabas F, Xie J, Serasinghe MN, et al. Mitochondrial metabolism in hematopoietic stem cells requires functional FOXO3. EMBO Rep. 2015;16(9):1164–76. https://doi.org/10.15252/embr.201439704.
•• Ho TT, Warr MR, Adelman ER, Lansinger OM, Flach J, Verovskaya EV, et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543(7644):205–10. https://doi.org/10.1038/nature21388. This study demonstrate that autophagy is necessary to preserve the regenerative capacity of old HSC by clearing active, healthy mitochondria and convincingly shows that about 30% of old HSC exhibit high autophagy and retain stemness like yong one.
Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208(3):455–67. https://doi.org/10.1084/jem.20101145.
Mortensen M, Watson AS, Simon AK. Lack of autophagy in the hematopoietic system leads to loss of hematopoietic stem cell function and dysregulated myeloid proliferation. Autophagy. 2011;7(9):1069–70.
Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nat Cell Biol. 2010;12(9):842–6. https://doi.org/10.1038/ncb0910-842.
Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494(7437):323–7. https://doi.org/10.1038/nature11895.
Ito K, Turcotte R, Cui J, Zimmerman SE, Pinho S, Mizoguchi T, et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science. 2016;354(6316):1156–60. https://doi.org/10.1126/science.aaf5530.
Kohler A, Schmithorst V, Filippi MD, Ryan MA, Daria D, Gunzer M, et al. Altered cellular dynamics and endosteal location of aged early hematopoietic progenitor cells revealed by time-lapse intravital imaging in long bones. Blood. 2009;114(2):290–8. https://doi.org/10.1182/blood-2008-12-195644.
Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299(5613):1751–3. https://doi.org/10.1126/science.1080418.
Erjavec N, Larsson L, Grantham J, Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21(19):2410–21. https://doi.org/10.1101/gad.439307.
Liu B, Larsson L, Caballero A, Hao X, Oling D, Grantham J, et al. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140(2):257–67. https://doi.org/10.1016/j.cell.2009.12.031.
Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454(7205):728–34. https://doi.org/10.1038/nature07212.
Sengupta A, Duran A, Ishikawa E, Florian MC, Dunn SK, Ficker AM, et al. Atypical protein kinase C (aPKCzeta and aPKClambda) is dispensable for mammalian hematopoietic stem cell activity and blood formation. Proc Natl Acad Sci U S A. 2011;108(24):9957–62. https://doi.org/10.1073/pnas.1103132108.
Wilson A, Ardiet DL, Saner C, Vilain N, Beermann F, Aguet M, et al. Normal hemopoiesis and lymphopoiesis in the combined absence of numb and numblike. J Immunol. 2007;178(11):6746–51.
•• Florian MC, Nattamai KJ, Dorr K, Marka G, Uberle B, Vas V, et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell aging. Nature. 2013;503(7476):392–6. https://doi.org/10.1038/nature12631. This paper highlights that Wnt5a treatment of young HSC induces aging-associated stem-cell apolarity, reduction of regenerative capacity and an aging-like myeloid-lymphoid differentiation skewing via activation of the small Rho GTPase Cdc42. They also identify that Wnt5a haploinsufficiency attenuates HSC aging, and stem-cell-intrinsic reduction of Wnt5a expression results in functionally rejuvenated aged HSC.
Beerman I, Bock C, Garrison BS, Smith ZD, Gu H, Meissner A, et al. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell. 2013;12(4):413–25. https://doi.org/10.1016/j.stem.2013.01.017.
Sun D, Luo M, Jeong M, Rodriguez B, Xia Z, Hannah R, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14(5):673–88. https://doi.org/10.1016/j.stem.2014.03.002.
Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23(8):413–8. https://doi.org/10.1016/j.tig.2007.05.008.
Han S, Brunet A. Histone methylation makes its mark on longevity. Trends Cell Biol. 2012;22(1):42–9. https://doi.org/10.1016/j.tcb.2011.11.001.
• Burman B, Zhang ZZ, Pegoraro G, Lieb JD, Misteli T. Histone modifications predispose genome regions to breakage and translocation. Genes Dev. 2015;29(13):1393–402. https://doi.org/10.1101/gad.262170.115. This article identifies specific histone modifications as facilitators of chromosome breakage and translocations using a combination of bioinformatics, biochemical analysis, and cell-based assays.
Burke B, Stewart CL. Functional architecture of the cell’s nucleus in development, aging, and disease. Curr Top Dev Biol. 2014;109:1–52. https://doi.org/10.1016/B978-0-12-397920-9.00006-8.
• Ugarte F, Sousae R, Cinquin B, Martin EW, Krietsch J, Sanchez G, et al. Progressive chromatin condensation and H3K9 methylation regulate the differentiation of embryonic and hematopoietic stem cells. Stem Cell Reports. 2015;5(5):728–40. https://doi.org/10.1016/j.stemcr.2015.09.009. This article identifies global chromatin rearrangements during stem cell differentiation and that heterochromatin formation by H3K9methylation regulates HSC differentiation.
Vas V, Senger K, Dorr K, Niebel A, Geiger H. Aging of the microenvironment influences clonality in hematopoiesis. PLoS One. 2012;7(8):e42080. https://doi.org/10.1371/journal.pone.0042080.
Vas V, Wandhoff C, Dorr K, Niebel A, Geiger H. Contribution of an aged microenvironment to aging-associated myeloproliferative disease. PLoS One. 2012;7(2):e31523. https://doi.org/10.1371/journal.pone.0031523.
Siclari VA, Zhu J, Akiyama K, Liu F, Zhang X, Chandra A, et al. Mesenchymal progenitors residing close to the bone surface are functionally distinct from those in the central bone marrow. Bone. 2013;53(2):575–86. https://doi.org/10.1016/j.bone.2012.12.013.
•• Kusumbe AP, Ramasamy SK, Itkin T, Mae MA, Langen UH, Betsholtz C, et al. Age-dependent modulation of vascular niches for haematopoietic stem cells. Nature. 2016;532(7599):380–4. https://doi.org/10.1038/nature17638. This article demonstrated the role of vascular niches and niche forming vessels in HSC aging, and highlighted the significance of endothelial cells Notch signalling in niche expansion.
Zhang Y, Depond M, He L, Foudi A, Kwarteng EO, Lauret E, et al. CXCR4/CXCL12 axis counteracts hematopoietic stem cell exhaustion through selective protection against oxidative stress. Sci Rep. 2016;6:37827. https://doi.org/10.1038/srep37827.
Tuljapurkar SR, McGuire TR, Brusnahan SK, Jackson JD, Garvin KL, Kessinger MA, et al. Changes in human bone marrow fat content associated with changes in hematopoietic stem cell numbers and cytokine levels with aging. J Anat. 2011;219(5):574–81. https://doi.org/10.1111/j.1469-7580.2011.01423.x.
Ergen AV, Boles NC, Goodell MA. Rantes/Ccl5 influences hematopoietic stem cell subtypes and causes myeloid skewing. Blood. 2012;119(11):2500–9. https://doi.org/10.1182/blood-2011-11-391730.
Chang KH, Sengupta A, Nayak RC, Duran A, Lee SJ, Pratt RG, et al. p62 is required for stem cell/progenitor retention through inhibition of IKK/NF-kappaB/Ccl4 signaling at the bone marrow macrophage-osteoblast niche. Cell Rep. 2014;9(6):2084–97. https://doi.org/10.1016/j.celrep.2014.11.031.
•• Guidi N, Sacma M, Standker L, Soller K, Marka G, Eiwen K, et al. Osteopontin attenuates aging-associated phenotypes of hematopoietic stem cells. EMBO J. 2017;36(7):840–53. https://doi.org/10.15252/embj.201694969. This paper convincingly shows that reduced expression of osteopoetin upon aging limits HSC regeneation, and identified thrombin-cleaved osteopoetin as a novel rejuvenation approach.
Quere R, Saint-Paul L, Carmignac V, Martin RZ, Chretien ML, Largeot A, et al. Tif1gamma regulates the TGF-beta1 receptor and promotes physiological aging of hematopoietic stem cells. Proc Natl Acad Sci U S A. 2014;111(29):10592–7. https://doi.org/10.1073/pnas.1405546111.
Blank U, Karlsson S. TGF-beta signaling in the control of hematopoietic stem cells. Blood. 2015;125(23):3542–50. https://doi.org/10.1182/blood-2014-12-618090.
Taniguchi Ishikawa E, Gonzalez-Nieto D, Ghiaur G, Dunn SK, Ficker AM, Murali B, et al. Connexin-43 prevents hematopoietic stem cell senescence through transfer of reactive oxygen species to bone marrow stromal cells. Proc Natl Acad Sci U S A. 2012;109(23):9071–6. https://doi.org/10.1073/pnas.1120358109.
Gonzalez-Nieto D, Li L, Kohler A, Ghiaur G, Ishikawa E, Sengupta A, et al. Connexin-43 in the osteogenic BM niche regulates its cellular composition and the bidirectional traffic of hematopoietic stem cells and progenitors. Blood. 2012;119(22):5144–54. https://doi.org/10.1182/blood-2011-07-368506.
Howcroft TK, Campisi J, Louis GB, Smith MT, Wise B, Wyss-Coray T, et al. The role of inflammation in age-related disease. Aging (Albany NY). 2013;5(1):84–93. https://doi.org/10.18632/aging.100531.
De Martinis M, Franceschi C, Monti D, Ginaldi L. Inflamm-aging and lifelong antigenic load as major determinants of aging rate and longevity. FEBS Lett. 2005;579(10):2035–9. https://doi.org/10.1016/j.febslet.2005.02.055.
Kovtonyuk LV, Fritsch K, Feng X, Manz MG, Takizawa H. Inflamm-aging of hematopoiesis, hematopoietic stem cells, and the bone marrow microenvironment. Front Immunol. 2016;7:502. https://doi.org/10.3389/fimmu.2016.00502.
Bersenev A, Rozenova K, Balcerek J, Jiang J, Wu C, Tong W. Lnk deficiency partially mitigates hematopoietic stem cell aging. Aging Cell. 2012;11(6):949–59. https://doi.org/10.1111/j.1474-9726.2012.00862.x.
Norddahl GL, Wahlestedt M, Gisler S, Sigvardsson M, Bryder D. Reduced repression of cytokine signaling ameliorates age-induced decline in hematopoietic stem cell function. Aging Cell. 2012;11(6):1128–31. https://doi.org/10.1111/j.1474-9726.2012.00863.x.
Pietras EM. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood. 2017;130(15):1693–8. https://doi.org/10.1182/blood-2017-06-780882.
• Josefsdottir KS, Baldridge MT, Kadmon CS, King KY. Antibiotics impair murine hematopoiesis by depleting the intestinal microbiota. Blood. 2017;129(6):729–39. https://doi.org/10.1182/blood-2016-03-708594. This paper demonstrated that microbiota sustain steady-state hematopoiesis through activation of Stat1 signaling.
Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15(3):374–81. https://doi.org/10.1016/j.chom.2014.02.006.
Heintz C, Mair W. You are what you host: microbiome modulation of the aging process. Cell. 2014;156(3):408–11. https://doi.org/10.1016/j.cell.2014.01.025.
•• Han B, Sivaramakrishnan P, Lin CJ, Neve IAA, He J, Tay LWR, et al. Microbial genetic composition tunes host longevity. Cell. 2017;169(7):1249–62 e13. https://doi.org/10.1016/j.cell.2017.05.036. This study highlighted the significance of bacterial genes on host aging and identified mTOR, JNK, Insulin signaling, and caloric restriction as a novel molecular targets.
Acknowledgments
The authors want to thank Ms. Margaret O’Leary for editing this manuscript.
The authors also want to thank the Cincinnati Children’s Hospital Medical Center and Hoxworth Blood Center for their continued support.
Funding
This study has been partly supported by the National Institutes of Health F31HL1324801 (M.J.A.), R01GM110628 (J.A.C), Leukemia and Lymphoma Society of North America (J.A.C.), and Williams Lawrence & Blanche Hughes Foundation (J.A.C.)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Abhishek K. Singh, Mark J. Althoff, and Jose A. Cancelas declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Role of Classical Signaling Pathways in Stem Cell Maintenance
Rights and permissions
About this article
Cite this article
Singh, A.K., Althoff, M.J. & Cancelas, J.A. Signaling Pathways Regulating Hematopoietic Stem Cell and Progenitor Aging. Curr Stem Cell Rep 4, 166–181 (2018). https://doi.org/10.1007/s40778-018-0128-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40778-018-0128-6