Abstract
Wheat powdery mildew is a severe disease affecting yield and quality. Host resistance was proved to be effective and environment-friendly. Wheat line Subtil is an elite germplasm resource resistant to 28 of 30 tested Bgt isolates. Genetic analysis showed that the powdery mildew resistance in Subtil was conferred by a single dominant gene, temporarily designated PmSub. Using bulked segregant analysis, PmSub was mapped to chromosome arm 5DS, and flanked by the markers Bwm16 and Cfd81/Bwm21 at 5.0 and 0.9 cM, respectively. Allelism tests further confirmed PmSub was allelic with documented Pm2 alleles. Then, homologous sequences of Pm2a related sequence was cloned from Subtil and Chinese Spring. It was completely identical to the reported Pm2a sequence, but significantly different from that of Chinese Spring. A marker SWGI067 was developed based on the sequence divergence of homologous sequence in Subtil and Chinese Spring. SWGI067 was closely linked to PmSub, indicating that the gene PmSub itself was different from the cloned Pm2a related sequence. Meanwhile, Subtil produced significantly different reaction pattern compared with other genotypes with Pm genes at or near Pm2 locus. Therefore, PmSub was most likely a new allele of Pm2. PmSub has opportunities for marker-assisted selecting for high-efficiency wheat improvement.
Similar content being viewed by others
Introduction
Wheat (Triticum aestivum L.) powdery mildew, caused by Blumeria graminis f. sp. tritici (Bgt), is one of the most damaging foliar diseases that occurs worldwide, especially with the deployment of dwarf and semi-dwarf cultivars and improvement of irrigation conditions1,2,3. Host resistance is proved to be an effective and safe method to minimize grain losses caused by the disease. However, resistance is often defeated by virulent mutants of the pathogen after long-term popularization of the cultivars with resistant gene(s)4,5. Previous studies indicated that most current wheat cultivars and breeding lines grown in China lacked effective resistance to powdery mildew (Pm)6. Therefore, it is urgent to identify more effective resistant sources among various germplasms to increase the genetic diversity of the resistant genes.
Up to now, 77 formally (Pm1-Pm54, Pm8 is allelic to Pm17, Pm18 = Pm1c, Pm22 = Pm1e, Pm23 = Pm4c, Pm31 = Pm21) and more than 30 temporarily designated (e.g. PmYB, PmWFJ, MlIw170) wheat Pm genes have been reported at 56 loci throughout all homoeologous chromosome groups7,8. Among these genes, there is an interesting multi-allelic phenomenon, that is, several Pm genes with different reaction patterns to Bgt isolates were located at the same locus in different genotypes. These loci include Pm1 (Pm1a-1e), Pm2 (2a-2c), Pm3 (3a-3j), Pm4 (4a-4d), Pm5 (5a-5e) and Pm24 (24a-24b) that were located at 7AL, 5DS, 1AS, 2AL, 7BL and 1DS, respectively7,9. This could due to the plant-pathogen interaction during long term deployment of the resistant cultivars or multiple generations of hybridization10,11. The new alleles may be very useful evolution, because when some alleles have lost effectiveness, new allelic variation may be present in other materials and provide broader resistant spectrum to different Bgt isolates, such as Pm2, Pm4 and Pm5 in several Chinese cultivars which increased the diversity of available resistance genes9,12,13.
Molecular markers are powerful tools for tagging genes and markerassisted selection (MAS)14. Using various kinds of markers, many favorable genes have been mapped to specific chromosomal loci7. In particular, with the development of high-throughput single nucleotide polymorphism (SNP) genotyping platforms based on wheat 9 K, 90 K and even 660 K SNP chips, high density linkage maps can be conducted using the SNP markers which can greatly increase the number of markers closely linked to targeted genes15,16,17,18. Using closely linked markers, the valuable genes can be rapidly transferred to other cultivars or pyramided with other desirable genes. For example, three QTLs conferring powdery mildew resistance were effectively selected in both greenhouse and field experiments19,20,21, and this increased the powdery mildew resistance in pyramided lines. We previously reported that the gene Pm2b was transferred to various susceptible cultivars, such as Shimai 15, Shixin 828, Gao 8901 etc., and efficiently selected by its closely linked markers to improve the powdery mildew resistance of the susceptible cultivars22. Apart from disease resistance, QTL/genes for some major economic traits, such as grain protein content and pre-harvest sprouting tolerance, have also been used for MAS in wheat breeding programs23,24,25,26.
In this study, the wheat line Subtil is highly resistant to 30 of Bgt isolates from different regions of China at the seedling stage in the greenhouse and immune to Bgt composite mixture at the adult stage in the filed of Shijiazhuang city of China. To make better use of this resistance resource, the following studies were carried out to: (1) determine the inheritance of powdery mildew resistance in Subtil; (2) map the resistance gene(s) in Subtil, and confirm the allelic relationship with the documented Pm genes; (3) compare reaction patterns to different Bgt isolates between Subtil and the genotypes carrying documented Pm genes; (4) distinguish PmSub with the cloned Pm2 sequence; and (5) investigate the applicability of closely linked markers for MAS.
Results
Inheritance of the powdery mildew resistance in Subtil
When inoculated with Bgt isolate E09, Subtil was immune with infection type (IT) 0, while Hengguan 35 was highly susceptible with IT 4. All the 25 F1 plants of Subtil × Hengguan 35 were immune with IT 0, in accord with that of the resistant parent, indicating the resistance gene in Subtil was dominant. Among the F2 population containing 162 plants, 119 were resistant with ITs 0–2; 43 were susceptible with ITs 3–4, fitting a single dominant gene segregation ratio (χ23:1 = 0.13, P = 0.72) (Table 1). The F2 population was then transplanted to the field, and 141 plants survived to produce F3 seeds. When tested with the same isolate, the F2:3 families segregated as 43 homozygous resistant (RR), 64 heterozygous resistant (Rr) and 34 were homozygous susceptible (rr), which confirmed single gene segregation ratio (χ21:2:1 = 2.35, P = 0.31). This gene was temporarily designated PmSub.
Molecular mapping of PmSub
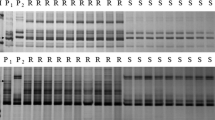
Initially, 310 SSR markers were surveyed their polymorphisms between parents Subtil and Hengguan 35, and the resistant and susceptible DNA bulks. Only the marker Cfd81 showed consistent polymorphism between the parents and bulks. Because Cfd81 was tightly linked to Pm227 and Pm4828, further 17 markers linked to Pm2 alleles or Pm48 were tested to survey the polymorphism between parents and bulks, including two SCAR markers Scar112 and Scar203, five SSR markers Gwm159, Cfd78, Wmc608, Cfd40 and Wmc805 and 10 SNP-derived SSR markers Bwm13, Bwm6, Bwm3, Bwm11, Bwm8, Bwm9, Bwm16, Bwm20, Bwm21 and Bwm25. Of these markers, 12 markers showed polymorphism between the parents and the bulks except for markers Bwm3, Bwm8, Bwm9, Bwm11 and Bwm13. Then all the 13 polymorphic markers containing Cfd81 were genotyped on the 141 F2:3 families of Subtil × Hengguan 35 (Fig. 1 & Fig. S1). Linkage analysis indicated that three markers Scar112, Gwm159 and Wmc805 were not linked to PmSub. A linkage map of PmSub was then constructed using the linked markers (Fig. 2). PmSub was flanked by the markers Cfd81/Bwm21 (distal) and Bwm16 (proximal) with genetic distances of 0.9 and 5.0 cM, respectively.
Examples of amplification patterns by the marker Bwm20 from selected F2:3 of Subtil × Hengguan 35. M: pUC19/MspI; PR: Subtil; PS: Hengguan 35; BR: Resistance bulked pool; BS: Susceptible bulked pool; R: Homozygous resistant F2:3 families; Rr: Heterozygous resistant F2:3 families; S: Homozygous susceptible F2:3 families. Black arrows indicated the polymorphic band (s).
The reaction patterns of Subtil and the lines with reported resistance alleles at or near the Pm2 locus
When inoculated with 30 Bgt isolates, Subtil was resistant to 28 of 30 isolates, while Ulka/*8 Cc (Pm2a), KM2939 (Pm2b), Niaomai (Pm2c), Tabasco (Pm48), D57-5D (PmD57-5D), Liangxing 66 (PmLX66), X3986-2 (PmX3986-2), Wanfengjian 34, Yingbo 700, Wennong 14, Zhongmai 155 and FG-1 were susceptible to six, three, three, four, six, seven, eleven, seven, one, seven, seven and six of the tested isolates, respectively (Table 1 & Fig. 3). Subtil showed a relatively broader resistant spectrum to different Bgt isolates, and its reaction pattern was different from those of the documented resistant stocks with Pm genes at/near the Pm2 locus. Thus, PmSub is most likely a new Pm gene.
Allelism of PmSub and the documented Pm genes on chromosome 5DS
To identify the allelic relationship between PmSub and the documented Pm genes on chromosome arm 5DS, F2 populations of the reciprocal crosses between Subtil (PmSub) and Ulka/*8 Cc (Pm2a), KM2939 (Pm2b), Niaomai (Pm2c), Liangxing 66 (PmLX66) were tested against Bgt isolate E09 avirulent to all these resistant stocks. All tested F2 plants of Subtil × genotypes with Pm2 alleles, including 10,318 plants of eight crosses, showed resistant to E09 (Table 2). Considering no susceptible plants were detected in these F2 populations, no recombination occurred between PmSub and Pm2 allelic loci. This indicated that PmSub was allelic with Pm2. Combined with the distinguishable reaction pattern to the different Bgt isolates, PmSub is most likely a new allele at the Pm2 locus. Furthermore, one of 1406 F2 plants of Subtil (PmSub) × Tabasco (Pm48) was susceptible to E09 with IT 4. This indicates that PmSub seems to be closely linked with Pm48.
Comparison of PmSub and the cloned Pm2 sequence
Using primers JS320 and JS305, a band was produced from PCR, and it was 4825 bp after Sanger sequencing. Then, using the 4825 bp DNA as template, the nested PCR was performed to obtain the first exon, which was a 3730 bp sequence in the amplified band (4083 bp) after Sanger sequencing. Meanwhile, a significant sequence difference was present in Chinese Spring using the same procedure, including a 12 bp insertion, a 13 bp deletion, a 33 bp deletion and many single nucleotide mutations. Compared with the cloned Pm2a, the first exon was completely same with that of Pm2a. The second and third exons were amplified by the primers JS350 and JS313. After Sanger sequencing, they were 58 and 46 bp bands respectively and also completely same with the second and third exons of Pm2a. Therefore, the cloned exons from Subtil were identical with those of Pm2a and significantly different from that of Chinese Spring.
To confirm the cloned sequence from Subtil was the gene PmSub itself that is indeed correlated with the powdery mildew resistance in Subtil, marker SWGI067 that can amplify part of the first exon was first used to test Subtil, Hengguan 35 and the resistant and susceptible bulks. It can amplify consistent polymorphism between parents and bulks. Then, SWGI067 was used to genotyped on the 141 F2:3 families of Subtil × Hengguan 35. Seven recombinants were detected in the F2:3 families, among which the genotypes of four segregated F2:3 families (No. 51, 54, 96 and 139) were homozygous susceptible as Hengguan 35, two segregated F2:3 families (No. 63 and 130) were homozygous resistant as Subtil and one homozygous resistant F2:3 family (No. 141) was heterozygous resistant. To avoid false hybrid strains of these recombinants, 50 SSR markers randomly distributed on 21 chromosomes of wheat were used to detect their genetic backgrounds. The results showed that the genetic backgrounds of these recombinants were accorded with their parents, and the recombinations were really existing. This demonstated that the first exon of the Pm2a related gene had certain genetic distance with PmSub. After calculation by Mapmarker 3.0, the genetic distance was 2.8 cM (distal) (Fig. 1), indicating that PmSub was likely different from the cloned sequence of Pm2a.
Potential of flanking markers for MAS
The flanking markers Cfd81 and Bwm16 of PmSub were assayed on 12 documented resistant stocks and 10 Chinese elite cultivars to investigate the potential of the markers for MAS. The polymorphic Cfd81 and Bwm16 alleles were present in Subtil and other Pm2 stocks, whereas not appeared in the 10 cultivars, indicating that when PmSub was transferred to these cultivars with no Pm genes at the Pm2 locus by conventional hybridization, the flanking markers Cfd81 and Bwm16 can be used in MAS (Fig. 4 & Fig. S2).
Amplification patterns of the marker Cfd81 on Subtil, documented stocks with Pm2 alleles and several wheat cultivars for validation of MAS. Lanes 1–25 are Subtil, Hengguan35, Resistance bulked pool, Susceptible bulked pool, Ulka/*8 Cc, KM2939, Niaomai, Brock, D57-5D, Liangxing 66, X3986-2, Zhongmai 155, Wennong 14, FG-1, Tabasco, Shi 4185, Shimai 15, Gao 8901, Han 6172, Han 7086, Kenong 9204, Jishi 02-1, Aikang 58, Baichun 5, Zhengmai 9023; M: pUC19/MspI; Black arrow indicates the polymorphic band in Subtil.
Discussion
Subtil is a highly resistant wheat breeding line in China. In five consecutive years, Subtil was tested against different Bgt isolates at seedling stage in the greenhouse, and mixed Bgt isolates collected from different regions of wheat production at adult stage in the field. Subtil consistently showed a stable, high level of resistance to powdery mildew. To identify the powdery mildew resistance, the segregation population of Subtil × Hengguan 35 was constructed for genetic analysis and molecular mapping of the Pm gene (s) in Subtil. A single dominant gene on chromosome arm 5DS, temporarily designated PmSub, was proved to provide the powdery mildew resistance in Subtil.
On chromosome arm 5DS, a series of Pm2 alleles (e.g. Pm2a, Pm2b, Pm2c, PmD57-5D, PmLX66 and PmX3986-2, etc) and Pm48 that closely linked to Pm2 have been reported22,28,29,30,31,32,33,34. Compared with the previously reported genes, PmSub showed relatively broad resistant spectrum, and was significantly different from the documented Pm genes on chromosome arm 5DS. Furthermore, allelism tests of not only between PmSub and Pm2 alleles but also PmSub and Pm48 were carried out to thoroughly confirm the allelic relationship with the documented Pm genes.
Recently, the Pm2a related sequence was cloned by mutant chromosome sequencing35. The homologous sequence was cloned in Subtil in this study. Although the homologous sequence in Subtil was same as the cloned Pm2a related sequence, it was closely linked but not co-segregated with PmSub, indicating that the cloned homologous sequence may not be the PmSub itself but a key factor of gene, and also be different from the cloned Pm2a related sequence. We presume that the reasons may be as follows: the cloned Pm2a sequence may not be the Pm2a gene itself, but a key factor in the upstream region of Pm2a. When this sequence is mutated, it will also lose the function. Although the homologous sequence in Subtil is same as the cloned Pm2a, it doesn’t mean that the PmSub sequence was same as the Pm2a sequence. More work need to be done in the future. For example, functional complemention verification by transgenosis need to be done to confirm if the function was obtained by transferring the cloned sequence of Pm2a; map-based cloning should be performed sequentially using forward genetic approaches to confirm the gene itself, etc.
In this study, a new allele joined the complex Pm2 allelic family. Like Pm1, Pm3, Pm4 and Pm5, and more and more new alleles with different resistant spectrum to multiple Bgt isolates were identified in Pm2 locus in recent years8,22,31,32,33,34. Previous studies indicated that part of the sequence variation may contribute to the phenomenon. For example, cloning of Pm3b10,36 demonstrated that about 3% sequence variation may result in different resistant spectrum of a series of Pm3 alleles. In the past, identification of new resistant resources mainly focus on the genes located at new loci7. With more and more resistance genes have been identified, exploring new alleles of the documented genes was also important for increasing the genetic diversity of the resistance genes. Even if the documented resistance genes in the commercial cultivars have lost or reduced resistance, their new allelic variations may have broader resistant spectrum to the virulent isolates, such as many new alleles of Pm2, Pm4 and Pm512,13,22,33,34,37. These allelic variations can increase the diversity at this locus, which will contribute to not only the genetic improvement of crops, but also the understanding of mechanism in the host-pathogen interactions38,39.
In wheat breeding, MAS is a rapid and effective way to transfer or pyramid excellent traits compared with conventional breeding methods. It has been used in many traits improvement successfully, such as disease resistance, adverse element tolerance and quality traits, etc40. In MAS, the key factor is the selection of applicable molecular markers41. In this study, Subtil is a wheat breeding line with superior agronomic performance. So, when the broad spectrum resistance gene PmSub was identified in Subtil, potential of flanking markers for MAS was investigated. Fortunately, two flanking markers have potential to detect PmSub in the tested cultivars with no Pm genes at or near Pm2 locus. Therefore, Subtil can be crossed to these cultivars, and their progenies can be high-efficiently selected by Cfd81 and Bwm16 for resistance breeding. Meanwhile, one other thing to note is the selecting marker Bwm16. Unlike Cfd81, it has a relatively far distance to PmSub, which will affect the efficiency and accuracy for MAS. So, more closely linked markers should be developed for MAS. Fine mapping and map-based cloning of PmSub will facilitate usefulness of this gene in wheat improvement.
Materials and Methods
Plant materials and Bgt isolates
Subtil is a winter wheat breeding line that is highly resistant to powdery mildew at both seedling and adult stages. Wheat cultivar Hengguan 35 is highly susceptible to powdery mildew and hence was used as susceptible parent in this study. An F2 population and 141 F2:3 families from the cross Subtil × Hengguan 35 were used to study the inheritance of powdery mildew resistance and map the resistance gene (s) in Subtil. The resistant stocks KM2939 (Pm2b)22, Niaomai (Pm2c)8, LiangXing 66 (PmLX66)31, X3986-2 (Pm3986-2)32, Wanfengjian 34 (PmWFJ)33; Yingbo 700 (PmYB)34, Wennong 14 (PmW14)42, Zhongmai 155 (PmZ155)3 and FG-1 (PmFG) are preserved in our lab. The resistant stock Ulka/*8 Cc (Pm2a)29, D57-5D (PmD57-5D)30 and German cultivar Tabasco (Pm 48)28 were provided by Prof. Hongyan Liu, Institute of Plant Protection, Henan Academy of Agricultural Sciences, Zhengzhou, Prof. Zhengqiang Ma in the Applied Plant Genomics Laboratory of Nanjing Agricultural University, Nanjing, and Prof. Shibin Cai, Institute of Food Crops, Jiangsu Academy of Agricultural Science, Nanjing, respectively. These resistant stocks were used in multi-isolates response comparisons with Subtil. Ten wheat cultivars in China were tested using molecular markers closely linked to the Pm gene in Subtil to validate the applicability of markers for MAS. Susceptible wheat cultivar Mingxian 169 was used as susceptible control for the test of powdery mildew resistance. Wheat cultivar Chinese Spring was used as the negative control which didn’t carry a Pm2 allele when homology-based cloning Pm2 alleles.
Twenty-eight single-pustule-derived powdery mildew virulent isolates were used to inoculate Subtil and the documented resistant stocks to test their reaction pattern to these Bgt isolates. These Bgt isolates have different virulence, and were kindly provided by Prof. Yilin Zhou, the State Key Laboratory for Biology of Plant Disease and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing. Bgt isolate E09 prevalent in North China was used to inoculate the mapping population of Subtil × Hengguan 35 for genetic analysis.
Phenotyping reactions to Bgt isolates
The reactions to Bgt isolates were tested in a greenhouse with a high humidity environment at 18 C/12 °C (day/ night) with a photoperiod of 12–14 h of light per day43. The Mingxian 169 seedlings were inoculated 30 Bgt isolates respectively and preserved separately in the glass tubes. The genotypes with documented Pm resistance alleles at/near Pm2 locus were grown in 128-well (3 cm × 3 cm) rectangular trays. Mingxian 169 was planted randomly in the trays as a susceptible check. These rectangular trays were prepared 30 copies. When the seedlings grown to one-leaf stage, every copy was inoculated a single Bgt isolate and preserved separately in a space. The F2 and F2:3 plants derived from the cross Subtil × Hengguan 35. Bgt isolate E09, avirulent on Subtil and virulent on Hengguan 35, was selected to inoculate Subtil, Hengguan 35 and their derived F1 hybrids, F2 and F2:3 populations (24 seedlings per family) of Subtil × Hengguan 35 for phenotypic survey and genetic analysis of the mapping populations. When the pustules were fully developed on the first leaf of Mingxian 169 at about 14–15 days after inoculation, infection types (ITs) for each plant were assessed on a 0–4 scale, and plants with ITs 0–2 were regarded as resistant and those with ITs 3 and 4 susceptible43.
Genotyping of the mapping population and map construction
Total genomic DNA of Subtil, Hengguan 35 and their derived F2:3 familes were separated from their young seedling leaves following the procedure of Ma et al.44. Resistant and susceptible DNA bulks were produced by mixing equal amounts of DNA of 10 homozygous resistant and 10 susceptible F2:3 families of Subtil × Hengguan 35 for bulked segregant analysis (BSA)45.
SSR markers evenly distributed across all 21 wheat chromosomes14,46,47 were selected to perform polymorphic marker survey between the parents and bulks. Sequence characterized amplified region (SCAR) markers SCAR203 and SCAR112, and SSR markers BWM3, BWM6, BWM8, BWM9, BWM11, BWM16, BWM20, BWM21 and BWM25 developed by Li et al.48 and Lu et al.49 respectively were also used to increase the marker density at the targeted interval. PCR was performed in a reaction volume of 10 ul containing 10–20 ng of template DNA, 2 pmol of each of the primers, 2 nmol of the dNTPs, 15 nmol of MgCl2, 0.1 U of Taq DNA polymerase, and 1 × PCR buffer. The PCR profile was one cycle of 94 °C for 3 min followed by 35 cycles of 94 °C for 30 s, 50–65 °C (depending on specific primers) for 40 s and 72 °C for 40 s, with a final extension at 72 °C for 5 min. PCR products were separated in 8% non-denaturing polyacrylamide gels (Acrylamide: Bisacrylamide = 25:1 or 39:1) with 1 × TBE buffer (90 mM tris-borate, 2 mM EDTA, PH 8.3), and visualized by silver straining50. Polymorphic markers were then genotyped on the F2:3 families of Subtil × Hengguan 35. After confirming the response genotypes through progeny testing of F2:3 families using the Bgt isolate E09, Chi-squared (χ2) tests were used to determine the goodness-of-fit of observed data with expected segregation ratios. Linkage analysis between polymorphic markers and the Pm gene in Subtil was performed by Mapmarker 3.0 with a LOD threshold score of 3.051. Genetic distances were estimated from recombination values using the Kosambi mapping function52.
Allelism test of the Pm gene (s) and the documented Pm genes on the same chromosome arm
After the Pm gene (s) in Subtil was mapped on chromosome arm 5DS, Subtil was crossed with the genotypes with documented Pm genes on the same chromosome arm, including Ulka/*8 Cc (Pm2a), KM2939 (Pm2b), Niaomai (Pm2c), Liangxing 66 (PmLX66) and Tabasco (Pm48). The F2 populations were inoculated with the Bgt isolate E09, which was avirulent to Subtil, Ulka/*8 Cc, KM2939, Niaomai, Liangxing 66 and Tabasco. From the ratio of resistant and susceptible numbers, the allelic relationships between the Pm gene(s) in Subtil and documented Pm genes were confirmed.
Molecular and genetic comparison of the Pm gene in Subtil with the cloned Pm2 sequence
Based on the recent report about cloning of a Pm2 allele Pm2a35, homologous sequence of PmSub was cloned using homology-based cloning. The first exon was firstly amplified using primers JS320 (Forward 5′-3′: ACGATGATGTGAATCTTCCGTG) and JS305 (Reverse 5′-3′: AATGATAGCATGCATTTGGAG). On this basis, the nested PCR was carried on to obtain the final sequence of the first exon using primers JS314 (Forward 5′-3′: TTTTCGCGGTATTGCTGGTG) and JS315 (Reverse 5′-3′: ACCTCCTGTCATCGGTTCAC). The second and third exons were obtained by JS350 (Forward 5′-3′: CCCTCCTCCTTGAAGAATCTGA) and JS313 (Reverse 5′-3′: GCACAAACTCTACCCTGTTCC). Then, the sequence of the PmSub were assembled and compared with the cloned sequence of Pm2a.
Based on the sequence divergence of the first exon in Subtil and Chinese Spring, a pair of primer SWGI067 was designed (Forward 5′-3′: CCTGGGAGGGCTCGGATCACTG, Reverse 5′-3′: GGAGGGATGAGCGGTTCTGTAG). The amplified sequence of SWGI067 can cover the diversity sequence interval of the first exon. Then, SWGI067 was used to genotype on the F2:3 familes of Subtil × Hengguan 35. If the cloned sequence of Subtil was indeed the gene PmSub itself, the marker SWGI067 will be co-segregated with the phenotype of F2:3 familes of Subtil × Hengguan 35, and if not, the gene PmSub itself may not be and different from the cloned sequence of Pm2a.
Validation of the closely linked markers in different genetic backgrounds
To evaluate the potential of the Pm gene(s) in Subtil for MAS, the flanking markers were tested against Subtil, 12 documented resistant stocks with Pm2 alleles and 10 Chinese wheat cultivars susceptible to powdery mildew. The patterns of the polymorphic bands were compared to assess the applicability of the markers in MAS. If polymorphic alleles were amplified between these wheat cultivars and Subtil, the markers can be used to detect the Pm gene (s) in Subtil when it was transferred to those cultivars by hybridization.
References
Bennett, F. G. A. Resistance to powdery mildew in wheat: A review of its use in agriculture and breeding programmes. Plant Pathol 33, 279–300 (1984).
Cowger, C. et al. Wheat powdery mildew. In:Sharma, I (ed) Disease resistance in wheat. CABI, Oxfordshire, 84–119 (2012).
Sun, H. G. et al. Resistance to powdery mildew in the wheat cultivar Zhongmai155: effectiveness and molecular detection of the resistance gene. Crop Sci 55, 1017–1025 (2015a).
Hsam, S.L.K., Zeller, F.J. Breeding for powdery mildew resistance in common wheat (Triticum aestivum L.). In:Belanger, R. R., Bushnell, W. R., Dik, A. J. & Carver, T. L. W. (eds) The powdery mildews, a comprehensive treatise. APS Press, St Paul MN 219–238 (2002).
Xiao, M. G. et al. Identification of the gene Pm47 on chromosome 7BS conferring resistance to powdery mildew in the Chinese wheat landrace Hongyanglazi. Theor Appl Genet 126, 1397–1403 (2013).
Li, H. J. et al. Response to powdery mildew and detection of resistance genes in wheat cultivars from China. Acta Agron Sin 37, 943–954 (2011).
McIntosh, R.A. et al. Catalogue of gene symbols for wheat: 2015–2016 supplement. http://www.shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp.
Xu, H. X. et al. Molecular tagging of a new broad-spectrum powdery mildew resistance allele Pm2c in Chinese wheat landrace Niaomai. Theor Appl Genet 128, 2077–2084 (2015).
Ma, P. T. et al. Characterization of a new Pm2 allele conferring powdery mildew resistance in the wheat germplasm line FG-1. Front Plant Sci 7, 546 (2016).
Srichumpa, P., Brunner, S., Keller, B. & Yahiaoui, N. Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol 139, 885–895 (2005).
Yahiaoui, N., Brunner, S. & Keller, B. Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J 47, 85–98 (2006).
Huang, X. Q., Wang, L. X., Xu, M. X. & Röder, M. S. Microsatellite mapping of the powdery mildew resistance gene Pm5e in common wheat (Triticum aestivum L.). Theor Appl Genet 106, 858–865 (2003).
Fu, B. S. et al. PmX: a recessive powdery mildew resistance gene at the Pm4 locus identified in wheat landrace Xiaohongpi. Theor Appl Genet 126, 913–921 (2013).
Somers, J. D., Isaac, P. & Edwards, K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109, 1105–1114 (2004).
Bérard, A. et al. High-throughput single nucleotide polymorphism genotyping in wheat (Triticum spp.). Plant Biotechnol J 7, 364–374 (2009).
Lai, K. T. et al. Single nucleotide polymorphism discovery from wheat next-generation sequence data. Plant Biotechnol J 10, 743–749 (2012).
Avni, R. et al. Ultra-dense genetic map of durum wheat × wild emmer wheat developed using the 90K iSelect SNP genotyping assay. Mol Breeding 34, 1549–1562 (2014).
Wang, S. C. et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12, 787–796 (2014).
Tucker, D. M., Griffey, C. A., Liu, S. & Saghai-Maroof, M. A. Potential for effective marker-assisted selection of three quantitative trait loci conferring adult plant resistance to powdery mildew in elite wheat breeding populations. Plant Breeding 125, 430–436 (2006).
Liu, J. et al. Molecular marker-facilitated pyramiding of different genes for powdery mildew resistance in wheat. Plant Breeding 119, 21–24 (2000).
Wang, X. Y., Chen, P. D. & Zhang, S. Z. Pyramiding and marker-assisted selection for powdery mildew resistance genes in common wheat. Acta Genet Sin 28, 640–646 (2001).
Ma, P. T. et al. Molecular mapping of a new powdery mildew resistance gene Pm2b in Chinese breeding line KM2939. Theor Appl Genet 128, 613–622 (2015a).
de Bustos, A., Rubio, P., Soler, C., Garcia, P. & Jouve, N. Marker assisted selection to improve HMW-glutenins in wheat. Euphytica 119, 69–73 (2001).
Davies, J., Berzonsky, W. A. & Leach, G. D. A comparison of marker-assisted and phenotypic selection for high grain protein content in spring wheat. Euphytica 152, 117–134 (2006).
Badea, A. et al. Phenotypic and marker-assisted evaluation of spring and winter wheat germplasm for resistance to fusarium head blight. Euphytica 164, 803–819 (2008).
Zhang, Z., Friesen, T. L., Simons, K. J., Xu, S. S. & Faris, J. D. Development, identification, and validation of markers for marker-assisted selection against the Stagonospora nodorum toxin sensitivity genes Tsn1 and Snn2 in wheat. Mol Breeding 23, 35–49 (2009).
Qiu, Y. C. et al. Identification of microsatellite markers linked to powdery mildew resistance gene Pm2 in wheat. Cereal Res Commun 34, 1267–1273 (2006).
Gao, H. D. et al. Genetic analysis and molecular mapping of a new powdery mildew resistance gene Pm46 in common wheat. Theor Appl Genet 125, 967–973 (2012).
McIntosh, R. A. & Baker, E. P. Cytogenetic studies in wheat IV Chromosomal location and linkage studies involving the Pm2 locus for powdery mildew resistance. Euphytica 19, 71–77 (1970).
Ma, H. Q. et al. Identification and mapping of a new powdery mildew resistance gene on chromosome 6D of common wheat. Theor Appl Genet 123, 1099–1106 (2011).
Huang, J. et al. Molecular detection of a gene effective against powdery mildew in the wheat cultivar Liangxing 66. Mol Breeding 30, 1737–1745 (2012).
Ma, P. T. et al. Inheritance and genetic mapping of a gene for seedling resistance to powdery mildew in wheat line X3986-2. Euphytica 200, 149–157 (2014).
Ma, P. T. et al. The gene PmWFJ is a new member of the complex Pm2 locus conferring unique powdery mildew resistance in wheat breeding line Wanfengjian 34. Mol Breeding 35, 210 (2015b).
Ma, P. T. et al. The gene PmYB confers broad-spectrum powdery mildew resistance in the multi-allelic Pm2 chromosome region of the Chinese wheat cultivar YingBo 700. Mol Breeding 35, 124 (2015c).
Sánchez-Martín, J. et al. Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol 17, 221 (2016).
Yahiaoui, N., Srichumpa, P., Dudler, R. & Keller, B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J 37, 528–538 (2004).
Schmolke, M., Mohler, V., Hartl, L., Zeller, F. J. & Hsam, S. L. K. A new powdery mildew resistance allele at the Pm4 wheat locus transferred from einkorn (Triticum monococcum). Mol Breeding 29, 449–456 (2012).
Prada, D. Molecular population genetics and agronomic alleles in seed banks: Searching for a needle in a haystack? J Exp Bot 60, 2541–2552 (2009).
Wicker, T. et al. The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat Genet 45, 1092–1096 (2013).
Gupta, P. K., Langridge, P. & Mir, R. R. Marker-assisted wheat breeding: present status and future possibilities. Mol Breeding 261, 145–161 (2010).
William, H. M., Trethowan, R. M. & Crosby-Galvan, E. M. Wheat breeding assisted by markers: CIMMYT’s experience. Euphytica 157, 307–319 (2007).
Song, W. et al. Chromosomal localization of the gene for resistance to powdery mildew in the wheat cultivar Wennong14. Acta Agron Sin 40, 798–804 (2014).
Si, Q. M., Zhang, X. X., Duan, X. Y., Sheng, B. Q. & Zhou, Y. L. On gene analysis and classification of powdery mildew (Erysiphe graminis f. sp. tritici) resistant wheat varieties. Acta Phytopathol Sin 22, 349–355 (1992).
Ma, Z. Q., Sorrells, M. E. & Tanksley, S. D. RFLP markers linked to powdery mildew resistance genes Pm1, Pm2, Pm3, and Pm4 in wheat. Genome 37, 871–875 (1994).
Michelmore, R. W., Paran, I. & Kesseli, R. V. Identification of markers linked to disease resistance gene by bulked segregant analysis: a rapid method to detect markers in specific genomic regions using segregating populations. Proc Natl Acad Sci USA 88, 9828–9832 (1991).
Röder, M. S. et al. A microsatellite map of wheat. Genetics 149, 2007–2023 (1998).
Paillard, S. et al. An integrative genetic linkage map of winter wheat (Triticum aestivum L.). Theor Appl Genet 107, 1235–1242 (2003).
Li, G. Q. et al. Molecular identification of a powdery mildew resistance gene from common wheat cultivar Brock (In Chinese). Acta Agron Sin 35, 1613–1619 (2009).
Lu, Y. Q. et al. Genetic mapping of a putative Agropyron cristatum-derived powdery mildew resistance gene by a combination of bulked segregant analysis and single nucleotide polymorphism array. Mol Breeding 35, 96 (2015).
Xu, H. X. et al. Identification and mapping pm2026, a recessive powdery mildew resistance gene in an einkorn (Triticum monococcum L.) accession. Theor Appl Genet 117, 471–477 (2008).
Lander, E. S. et al. Map marker: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181 (1987).
Kosambi, D. D. The estimation of map distance from recombination values. Ann Eugen 12, 172–175 (1943).
Acknowledgements
This research was financially supported by the National Key Research and Development Program of China (No. 2016YFD0102002), the National Natural Science Foundation of China (No. 31771793; No. 31671771) and the Chinese Academy of Sciences (No. XDA08030107).
Author information
Authors and Affiliations
Contributions
H. Xu, P. Ma and D. An designed the study, X. Fu and X. Zhang collected the plant materials, H. Xu constructed the populations, Y. Jin, P. Ma, L. Song and Y. Xu performed the experiments, H. Xu and Y. Jin analyzed the data, Y. Jin wrote the draft manuscript, H. Xu, P. Ma and D. An revised the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jin, Y., Xu, H., Ma, P. et al. Characterization of a new Pm2 allele associated with broad-spectrum powdery mildew resistance in wheat line Subtil. Sci Rep 8, 475 (2018). https://doi.org/10.1038/s41598-017-18827-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-18827-4
- Springer Nature Limited