Abstract
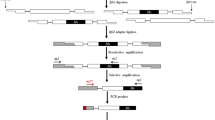
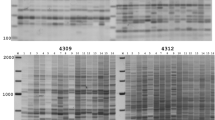
Variation is the primary source for plant and animal breeding and evolution, and hence, the detection of variation is an integral part of genetics, breeding, and ecology. Various molecular marker systems have been developed to detect genetic variation. They detect sequence variations (e.g., restriction fragment length polymorphism, randomly amplified polymorphic DNA, amplified fragment length polymorphism) or simple sequence motif variations. However, a large portion of genomic variation is derived from the transposition of transposable elements (TEs), which are major denizens of most eukaryote genomes. Therefore, molecular markers derived from TEs are valuable resources for dissecting genomes in plants and animals. Because class I retrotransposons transpose by a “copy-and-paste” semi-conservative manner, retrotransposon-based markers (e.g., Inter-retrotransposon amplified polymorphism, retrotransposon-microsatellite amplified polymorphism, sequence-specific amplified polymorphism) can reveal highly accurate phylogenetic relationships among related taxa as well as among accessions within a species. Transposon display based on class II DNA transposons has also been used in various genetics fields. A large amount of fairly accurate genome sequences are now being generated, and computational biology allows us to mine the TEs on a genome-wide scale. Thus, TE-based molecular markers are adding another venue to the other marker systems used for the molecular dissection of genomes.




Similar content being viewed by others
References
Bachmann K (1994) Molecular markers in plant ecology. New Phytol 126:403–418
Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meely RB, Briggs SP (1995) Cloning and characterization of the maize An1 gene. Plant Cell 7:75–84
Bonchev G, Parisod C (2013) Transposable elements and microevolutionary changes in natural populations. Mol Ecol Res 13:765–775
Borevitz JO, Liang D, Plouffe D, Chang HS, Zhu T, Weigel D, Berry CC, Winzeler E, Chory J (2003) Large-scale identification of single-feature polymorphism in complex genome. Genome Res 13:513–523
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Branco CJ, Vieira EA, Malone G, Kopp MM, Malone E, Bernardes A, Mistura CC, Carvalho FIF, Oliveria CA (2007) IRAP and REMAP assessment of genetic similarity in rice. J Appl Genet 48:107–113
Bureau T, Wessler SR (1992) Tourist: a large family of inverted-repeat element frequently associated with maize genes. Plant Cell 4:1248–1283
Bureau T, Wessler SR (1994) Stowaway: a large-family of invert-repeat element family frequently associated with maize genes. Plant Cell 4:1283–1294
Chagné D, Gasic K, Crowhurst RN, Han Y, Bassett HC, Bowatte DR, Lawrence TJ, Rikkerink EHA, Gardiner SE, Korban SS (2008) Development of a set of SNP markers present in expressed genes of the apple. Genomics 92:353–358
Craig NL, Craigie R, Gellert M, Lambowitz AM (2002) Mobile DNA II. Am Soc Microbiol Press, Washington
Das L, Martienssen R (1995) Site-selected transposon mutagenesis at the hcf106 locus in maize. Plant Cell 7:287–294
Du C, Fefelova N, Caronna J, He L, Dooner HK (2009) The polychromatic Helitron landscape of the maize genome. Proc Natl Acad Sci USA 106:1191–1192
Edwards M (2013) Whole-genome sequencing for marker discovery. In: Henry RJ (ed) Molecular markers in plants. Wiley-Blackwell Pub, Ames, pp 21–34
Ellergren H (2004) Microsatellites: simple sequence with complex evolution. Nat Rev Genet 5:435–445
Fan F, Cui B, Zhang T, Ding G, Wen X (2014) LTR-retrotransposon activation, IRAP marker development and its potential in genetic diversity assessment of masson pine (Linus massoniana). Tree Genet Genomics 10:2013–2222
Fedoroff NV (2013) The discovery of transposition. In: Fedoroff NV (ed) Plant transposons and genome dynamics in evolution. Wiley-Blackwell Inc, Iowa, pp 3–14
Fedoroff NV, Bennetzen JL (2013) Transposon, genomic shock, and genome expansion. In: Fedoroff NV (ed) Plant transposons and genome dynamics in evolution. Wiley-Blackwell Inc, Iowa, pp 181–201
Feschotte C, Zhang X, Wessler SR (2002a) Miniature inverted-repeat transposable elements and their relationship to established DNA transposons. In: Craig NL, Cragie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. ASM Press, Washington, pp 1147–1158
Feschotte C, Jiang N, Wessler SR (2002b) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3:329–341
Finnegan DJ (1989) Eukaryotic transposable elements and genome evolution. Trends Genet 5:103–107
Ganal MW, Altman T, Röder MS (2009) SNP identification in crop plants. Curr Opin Plant Biol 12:211–217
Ganapathy PS, Scandalios JG (1973) Malate dehydrogenase isozymes in haploid and diploid Datura species. Their use as markers in somatic cell genetics. J Hered 64:186–188
Gore MA, Wright MH, Ersoz ES, Bouffard P, Szekeres ES, Jarvie TP, Hurwitz BL, Narechania A, Harkins TT, Grills GS et al (2009) Large-scale discovery of gene enriched SNPs. Plant Genome 2:121–133
Grzebelus D (2006) Transposon insertion polymorphism as a new source of molecular markers. J Fruit Ornam Plant Res 14:21–29
Henry RJ (2013) Evolution of DNA marker technology in plants. In: Henry RJ (ed) Molecular markers in plants. Wiley-Blackwell Pub, Ames, pp 3–20
Huang X, Lu G, Zhao Q, Liu X, Han B (2008) Genome-wide analysis of transposon insertion polymorphisms reveals intraspecific variation in cultivated rice. Plant Physiol 148:25–40
Hubby JL, Lewontin RC (1966) A molecular approach to the study of genic heterozygosity in natural populations, I. The number of alleles at different loci in Drosophila pseudoobscura. Genetics 54:577–594
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity arrays: a solid state technology for sequence information independent genotyping. Nucl Acids Res 29(4):e25
Jiang GL (2013) Moleular markers and marker-assisted breeding in plants. In: Andersen SB (ed) Agricultural and biological science. Plant Breeding from Laboratories to Fields. doi:10.5772/52583
Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR (2003) An active DNA transposon family in rice. Nature 421:163–167
Jing R, Vershinin A, Grzebyta J, Smýkal P, Marsshall D, Ambrose MJ, Ellis TN, Flavell AJ (2010) The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymoprphism (RIBP) marker analysis. BMC Evol Biol 10:44
Josefesson C, Dilkes B, Comai L (2006) Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol 16:1322–1328
Kalendar R, Schulman AH (2006) IRAP and REMAP for retrotransposon-based genotyping and fingerprinting. Nat Protocols 1:2478–2484
Kalendar R, Grob T, Regina M, Suoniemi A, Schulman AH (1999) IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor Appl Genet 98:704–711
Kalendar R, Flavell AJ, Ellis T, Sjakste T, Moisy C, Schulman A (2011) Analysis of plant diversity with retrotransposon-based molecular markers. Heredity 106:520–530
Kapitonov V, Jurka J (2001) Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci USA 98:8714–8719
Kashkush K, Feldman M, Levy AA (2002) Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160:1651–1659
Kelly LJ, Leitch IJ (2011) Exploring giant plant genomes with next-generation sequencing technology. Chromosome Res 19:939–953
Kikuchi K, Terauchi K, Wada M, Hirano HY (2003) The plant MITE mPing is mobilized in anther culture. Nature 421:167–170
Kim H, Terakami S, Nishitani C, Kurita K, Kanamori H, Katayose Y, Sawamura Y, Saito T, Yamamoto T (2012) Development of cultivar-specific DNA markers based on retrotransposon-based insertional polymorphism in Japanese pear. Breed Sci 62:53–62
Korswagen HC, Durbin RM, Smits MT, Plasterk RHA (1996) Transposon TC1-derived, sequence-tagged sites in Caenorhabiditis elegans as markers for genetic mapping. Proc Natl Acad Sci USA 93:14680–14685
Kumar A, Hirochika H (2001) Applications of retrotransposons as genetic tools in plant biology. Trends Plant Biol 6:127–134
Kumar R, Qiu J, Joshi T, Valliyodan B, Xu D, Nguyen HT (2007) Single feature polymorphism discovery in rice. PLoS One 2:e284
Kunze R, Weil CF (2002) hAT and CACTA superfamilies’ in plant genomes. ASM Press, Washington, pp 565–612
Kwon SJ, Park KC, Kim JH, Lee JK, Kim NS (2005) Rim 2/Hipa CACTA transposon display: a new genetic marker technique in Oryza species. BMC Genet 6:15
Kwon SJ, Lee JK, Hong SW, Park YJ, McNally KL, Kim NS (2006a) Genetic diversity and phylogenetic relationship in AA Oryza species as revealed by Rim 2/Hipa CACTA transposon display. Genes Gent Syst 81:93–101
Kwon SJ, Hong SW, Son JH, Lee JK, Cha YS, Eun MY, Kim NS (2006b) CACTA and MITE transposon distribution in a recombinant genetic map of rice. Mol Cells 21:360–366
Le QH, Bureau T (2004) Prediction and quality assessment of transposon insertion display data. Biotechniques 36:222–228
Lee SI, Kim NS (2014) Transposable elements and genome size variations in plants. Genomics Inform 12:87–97
Lee JK, Park JY, Choi SH, Kim JH, Choi JK, Min HK, Park CH, Kim NS (2004) Genetic mapping of maize with the intermated Mo17 × KW7 population using MITE-AFLP and SSR markers. Kor J Genet 26:63–72
Lee SI, Park KC, Ha MW, Kim KS, Jang YS, Kim NS (2012) CACTA transposon-derived Ti-SCARs for cultivar fingerprinting in rapeseed. Genes Genom 34:575–579
Leeton PR, Smyth DR (1993) An abundant LINE-like elements amplified in the genome of Lilium speciosum. Mol General Genet 237:97–104
Levin H (2002) Newly identified retrotranspoons of the Ty3/gypsy class in fungi, plants, and vertebrates. In: Craig NL, Cragie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. ASM Press, Washington, pp 684–701
Liu B, Wendel JF (2000) Retrotransposon activation followed by rapid repression in introgressed rice plants. Genome 43: 874–880. In: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) Mobile elements II. American Society of Microbiology Press, Washington, DC
Lockton S, Ross-Ibarra J, Gaut BS (2008) Demography and weak selection drive patterns of transposable element diversity in natural Arabidopsis lyrata. Proc Nat Acad Sci USA 105:13965–13970
Mammadov J, Aggarwal T, Buyarapu R, Kumpatala S (2012) SNP markers and their impact on plant breeding. Int J Plant Genomics. doi:10.1155/2012/728398
Martienssen RA, Chandler VL (2013) Molecular mechanisms of transposon epigenetic regulation. In: Fedorof NV (ed) Transposons and genome dynamics in evolution. Wiley-Blackwell Pub, Ames, pp 71–92
McClintock B (1954) Mutations in maize and chromosomal abberrations in Neurospora. Carnegie Inst 53:254–260 Wanshingtonm Yearbook
McClintock B (1984) The significance of responses of the genome to challenge. Science 226:792–801
Moisy C, Garrison KE, Meredith CP, Pelsy F (2008) Characterization of ten novel Ty1/copia-like retrotransposon families of the grapevine genome. BMC Genom 9:469
Monden Y, Naito K, Okumoto Y, Saito H, Oki N, Tsukiyama T, Ideta O, Nakazaki T, Wessler SR, Tanjisaka T (2009) High potential of a transposon mPing as a marker system in japonica × japonica cross in rice. DNA Res 16:131–140
Moran JV, Gilbert N (2002) Mammalian LINE-1 retrotransposons and related elements. In: Craig NL, Cragie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. ASM Press, Washington, pp 836–869
Oliver KR, Greene WK (2009) Transposable elements: powerful facilitators of evolution. BioEssays 31:703–714
Oliver KR, McComb JA, Greene W (2013) Transposable elements: powerful contributors to angiosperm evolution and diversity. Mol Biol Evol 5:1886–1901
Parisod C, Senerchia N (2012) Responses of transposable elements to polyploidy. In: Grandbastien M-A, Casacuberta J (eds) Plant transposable elements. Topics in current genetics, vol 24. Springer, Heidelberg, pp 147–168
Parisod C, Salmon A, Zerjal T, Tenaillon M, Grandbastien M-A, Ainouche M (2009) Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol 184:1003–1015
Park KC, Kim NS (2012) Differentiation of CACTA-like elements in Arabidopsis. In: Pontarotti P (ed) Evolutionary biology: mechanism and trend. Springer, Marseille, pp 325–342
Park KC, Chung CS, Song MT, Kim NS (2003a) A new MITE family, Pangrangja, in Gramineae species. Mol Cells 15:373–380
Park KC, Kim NH, Cho YS, Kang KH, Lee JK, Kim NS (2003b) Genetic variations of AA genome Oryza species measured by MITE-AFLP. Theor Appl Genet 107:203–209
Park KC, Lee JK, Kim NH, Shin YB, Lee JH, Kim NS (2003c) Genetic relationship and genetic variation in Oryza species using MITE-AFLP. Genes Genet System 78:235–243
Park Y-J, Lee JK, Kim N-S (2009) Simple sequence repeat polymorphisms (SSRPs) for evaluation of molecular diversity and germplasm classification of minor crops. Molecules 14:4546–4569
Park KC, Park NI, Lee SI, Kim KS, Chang YS, Kim N-S (2014) A new active CACTA element and transposition activity in ecotype differentiation of Arabidopsis. Genes Genom 36:229–236
Paux E, Faure S, Choulet F, Roger D, Gauthier V, Martinant JP et al (2010) Insertion site-based polymorphism markers open new perspectives for genome saturation and marker assisted selection in wheat. Plant Biotech J 8:196–210
Peterson PA (1953) A multiple pale green locus in maize. Genetics 38:682–683
Petit M, Guidat C, Daniel J et al (2010) Mobilization of retrotransposons in synthetic allotetraploid tobacco. New Phytol 186:135–137
Poczai P, Varga I, Laos M, Cseh A, Bell N, Valkonen JPT, Hyvönen J (2013) Advances in plant gene-targeted and functional markers: a review. Plant Methods 9:6
Prince JP, Tanksley SD (1992) Restriction fragment length polymorphism in plant breeding and genetics. Proceed R Soc Edinb Sect B Biol Sci 99:23–29
Rowold DJ, Herrara RJ (2000) Alu element and the human genome. Genetica 108:57–72
Sakamoto K, Okada N (1985) Rodent type 2 Alu family, rat identifier sequence, rabbit C family, and bovine or goat 73-bp repeat may have evolved from tRNA genes. J Mol Evol 22:134–140
Sanchez AC, Brar DS, Huang N, Li Z, Khush GS (1999) Sequence tagged site marker-assisted selection for three bacterial blight resistance genes in rice. Crop Sci 40:792–797
Schulman AH (2007) Molecular markers to assess genetic diversity. Euphytica 158:313–321
Schulman AH, Wicker T (2013) A field guide to transposable elements. In: Fedoroff NV (ed) Plant transposons and genome dynamics in evolution. Wiley Blackwell, Oxford, pp 15–40
Singer T, Fan Y, Chang HS, Zhu T, Hazen SP, Briggs SP (2006) A high resolution map of Arabidospis recombinant inbred lines by whole genome exon-array hybridization. PLoS Genet 2:e144
Syed NH, Flavell AJ (2007) Sequence specific amplification polymorphisms (SSAP): a multiple-locus approach for analyzing transposon insertions. Nat Protocols 1:2746–2752
Talbert LE, Blake NK, Chee PW, Blake TK, Magyar GM (1994) Evaluation of “sequence-tagged-site” PCR products as molecular markers in wheat. Theor Appl Genet 87:789–794
Tam SM, Mhiri C, Vogelaar A, Kerkveld M, Pearce SR, Grandbastien MA (2005) Comparative analyses of genetic diversities within tomato and pepper collections detected by retrotransposon-based SSAP, AFLP, and SSR. Theor Appl Genet 110:819–831
Tanksley SD (1983) Molecular markers in plant breeding. Plant Mol Biol Rep 1:3–8
Tanksley SD, Young ND, Paterson AH, Bonierbale MW (1989) RFLP mapping in plant breeding: new tools for an old science. Nat Biotechnol 7:257–264
Tauts D, Renz M (1984) Simple sequence are ubiquitous repetitive components of eukaryotic genomes. Nucl Acids Res 12:4127–4138
Temnykh S, DeClerk G, Lukashova A, Lipovich L, Cartihour S, McCouch S (2001) Computational and experimental analysis microsatellite in rice (Oryza sativa L.): frequency, length variation, transposon association, and genetic marker potential. Genome Res 11:1441–1452
Thomas MR, Scott NS (1993) Microsatellite repeats in grapevine reveal DNA polymorphism when analyzed as sequence-tagged sites (STSs). Theor Appl Genet 86:985–990
Van den Broeck D, Maes T, Sauer M, Zethof J, De Keukelerie P, D’hauw M, Van Montagu M, Gerats T (1998) Transposon display identifies individual transposable elements in high copy number lines. Plant J 13:121–129
Varshney RK, Nayak SN, May GD, Jackson SA (2009) Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol 27:522–529
Vitte C, Ishii T, Lamy F, Brar D, Panaud O (2004) Genomic paleontology provides evidence for two distinct origins of Asian rice (Oryza sativa L.). Mol Gen Genomics 272:504–511
Vos P, Hogers R, Bleeker M, Reijans M, van der Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucl Acids Res 21:4407–4414
Voytas DF, Boeke JD (2002) Ty1 and Ty5 of Saccaromyces cereviceae. In: Craig NL, Cragie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. ASM Press, Washington, pp 631–662
Vukich M, Schulman AH, Giordani T, Natali I, Kalendar R, Cavallini (2009) Genetic variability in sunflower (Helianthus annus L.) and in the Helianthus genus as assessed by retrotransposon-based molecular markers. Theor Appl Genet 11:1027–1038
Wicker T, Guyot R, Yahiaouri M, Keller B (2003) CACTA transposons in Triticeae. A diverse family of high-copy repetitive elements. Plant Physiol 132:52–63
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Willams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl Acids Res 18:6531–6535
Woodrow P, Pontecorvo G, Fantaccione S, Fuggi A, Kafantaris I, Parisi D, Carillo P (2010) Polymorphism of a new Ty1-copia retrotransposon in durum wheat under salt and light stresses. Theor Appl Genet 121:311–322
Xing J, Witherspoon DJ, Jorde LB (2013) Mobile element biology: new possibilities with high-throughput sequencing. Trends Genet 29:280–289
Zhang X, Wessler SR (2004) Genome-wide comparative analysis of the transposable elements in the related species Arabidopsis thaliana and Brassica oleracea. Proc Natl Acad Sci USA 101:5589–5594
Zhuang J, Wang J, Theurkarf W, Weng Z (2014) TEMP: a computational method for analyzing transposable element polymorphism in populations. Nucl Acids Res. doi:10.1093/nar/gku323
Acknowledgments
This work was funded by a grant from Golden Seed Project, Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development of Korea (RDA), and Korea Forest Service (KFS).
Conflict of interest
Authors declare no conflict of interest on the contents of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Roy, N.S., Choi, JY., Lee, SI. et al. Marker utility of transposable elements for plant genetics, breeding, and ecology: a review. Genes Genom 37, 141–151 (2015). https://doi.org/10.1007/s13258-014-0252-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-014-0252-3