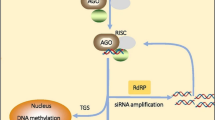
Abstract
RNA sequencing technology has revealed a vast number of small RNAs (sRNAs) involved in plant development, stress management, and crop improvement. With improvements in molecular biology techniques and bioinformatic tools, sRNAs and their regulations have taken a mainstream position in various research fields, including plant science. Moreover, recent findings emphasize the significance of ncRNAs and sRNAs in spliceosome machinery. Non-coding RNAs in plants consist of ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), microRNAs (miRNAs) (19–25 nt), short interfering RNAs (21–23 nt), and long non-coding RNAs (lncRNAs) (over 200 nt). The miRNAs and siRNAs have similar sizes but originate from structurally different RNA molecules and follow distinct biogenesis pathways with different modes of action. Uncovering their impact on plant developmental patterns and stress regulatory mechanisms assumed paramount importance in plant stress biology, specifically projects focused on crop improvement. Plants constantly face and adapt to environmental stresses that hinder their growth and development. Adverse effects of abiotic stresses, such as drought, heat, cold, and salinity, on plant productivity are well documented. With the onset of environmental stresses, certain sRNAs and ncRNAs take up prominent roles in cellular homeostasis and coordinate the stress responses. This review explores the functions of sRNAs and their interactions during plant development and stress regulation. The theme of this paper becomes pertinent given the challenge of developing crops for future environments.
Graphical abstract











Similar content being viewed by others
References
Achard P, et al. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–65.
Ahmed W, Xia Y, Zhang H, Li R, Bai G, Siddique KHM, et al. Identification of conserved and novel miRNAs responsive to heat stress in flowering Chinese cabbage using high-throughput sequencing. Sci Rep. 2019;9:14922. https://doi.org/10.1038/s41598-019-51443-y.
Akdogan G, Tufekci ED, Uranbey S, Unver T. miRNA-based drought regulation in wheat. Funct Integr Genomics. 2016;16(3):221–33.
Allen RS, et al. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci U S A. 2007;104:16371–6.
Arribas-Hernández L, Marchais A, Poulsen C, Haase B, Hauptmann J, Benes V, et al. The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. Plant Cell. 2016;28:1563–80. https://doi.org/10.1105/tpc.16.00121.
Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2- like target genes. Plant Cell. 2003;15:2730–41.
Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ. pho2, a phosphate over accumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 2006;141:1000–11.
Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev PlantBiol. 2013;64:137–59. https://doi.org/10.1146/annurev-arplant-050312120043.
Barakat A, Sriram A, Park J, Zhebentyayeva T, Main D, Abbott A. Genome wide identification of chilling responsive microRNAs in Prunus persica. BMC Genom. 2012;13:1–11.
Bartel DP. MicroRNAs: genomics, biogenesis mechanism, and function. Cell. 2004;116:281–97. https://doi.org/10.1016/S0092-8674(04)00045-5.
Barrera-Figueroa BE, Gao L, Wu Z, Zhou X, Zhu J, Jin H, Liu R, Zhu J-K. High throughput sequencing reveals novel and abiotic stress-regulated microRNAs in the inflorescences of rice. BMC Plant Biol. 2012;12:1–11.
Berger Y, et al. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development. 2009;136(823–832):49.
Bernstein E, Caudy AA, Hammond SM. Hannon GJ Rolefor a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–6. https://doi.org/10.1038/35053110.
Bian H, Xie Y, Guo F, Han N, Ma S, Zeng Z, Wang J, Yang Y, Zhu M. Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 2012;196:149–61.
Bian XB, Yu PC, Dong L, Zhao Y, Yang H, Han YZ, Zhang LX. Regulatory role of non-coding RNA in ginseng rusty root symptom tissue. Sci Rep. 2021;11:9211.
Blein T, et al. A conserved molecular framework for compound leaf development. Science. 2008;322:1835–9.
Bhutia KL, Khanna VK, Meetei TNG, Bhutia ND. Effects of climate change on growth and development of chilli. Agrotechnology. 2018;7(2):1–4.
Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–91.
Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. Anantagonistic function for Arabidopsis DCL2 in development and a newfunc- tion for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–56. https://doi.org/10.1038/sj.emboj.7601217.
Brant E, Budak H. Plant small non-coding RNAs and their roles in biotic stresses. Front Plant Sci. 2018;9:1038. https://doi.org/10.3389/fpls.2018.01038.
Budak H, Zhang B. MicroRNAs in model and complex organisms. Funct Integr Genomics. 2017;17:121–4. https://doi.org/10.1007/s10142-017-0544-1.
Busch BL, et al. Shoot branching and leaf dissection in tomato are regulated by homologous gene modules. Plant Cell. 2011;23:3595–609.
Cagirici HB, Alptekin B, Budak H. RNA sequencing and co-expressed long non-coding RNA in modern and wild wheats. Sci Rep. 2017;7:10670.
Cartolano M, et al. A conserved microRNA module exerts homeotic control over Petunia hybrida and Antirrhinum majus floral organ identity. Nat Genet. 2007;39:901–5.
Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94.
Chen X. Small RNAs and their roles in plant development. BioloAnnu Rev Cell Dev Biol. 2009;25:21–44.
Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–5.
Chen Z, Hu L, Han N, Hu J, Yang Y, Xiang T, Zhang X, Wang L. Overexpression of a miR393-resistant form of transport inhibitor response protein 1 (mTIR1) enhances salt tolerance by increased osmoregulation and Na+ exclusion in Arabidopsis thaliana. Plant Cell Physiol. 2015;56:73–83.
Chitwood DH, Timmermans MCP. Small RNAs are on the move. Nature. 2010;467:415–9.
Chuck G, et al. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet. 2007;39:1517–21.
Chuck G, et al. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development. 2010;137:1243–50.
Contreras-Cubas C, Palomar M, Arteaga-Vazquez M, Reyes JL, Covarrubias AA. Non-coding RNAs in the plant response to abiotic stress. Planta. 2012;236:943–58.
D’Ario M, Griffiths-Jones S, Kim M. Small RNAs: big impact on plant development. Trends Plant Sci. 2017;22(12):1056.
Das A, Saxena S, Kumar K, Tribhuvan KU, Singh NK, Gaikwad K. Non-coding RNAs having strong positive interaction with mRNAs reveal their regulatory nature during flowering in a wild relative of pigeonpea (Cajanus scarabaeoides). Mol Biol Rep. 2020;47:3305–17.
Ding Y, Ma Y, Liu N, Xu J, Hu Q, Li Y, et al. microRNAs involved in auxin signalling modulate male sterility under high-temperature stress in cotton (Gossypium hirsutum). Plant J. 2017;91:977–94. https://doi.org/10.1111/tpj.13620.
Djami-Tchatchou AT, Sanan-Mishra N, Ntushelo K, Dubery IA. Functional roles of microRNAs in agronomically important plants—potential as targets for crop improvement and protection. Front Plant Sci. 2017;8:378. https://doi.org/10.3389/fpls.2017.00378.
Esmaeli F, Shiran B, Fallahi H, Mirakhorli N, Budak H, Martínez-Gómez P. In silico search and biological validation of microRNAs related to drought response in peach and almond. Funct Integr Genomics. 2017;17:189–201.
Feldmann KA. Steroid regulation improves crop yield. Nat Biotechnol. 2006;4:46–7. https://doi.org/10.1038/nbt0106-46.
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11.
Gahlaut V, Baranwal VK, Khurana P. miRNomes involved in imparting thermotolerance to crop plants. 3 Biotech. 2018;8:497. https://doi.org/10.1007/s13205-018-1521-7.
Gaillochet C, Lohmann JU. The never-ending story: from pluripotency to plant developmental plasticity. Development. 2015;142:2237–49.
Gao P, Bai X, Yang L, Lv D, Pan X, Li Y, Cai H, Ji W, Chen Q, Zhu Y. osa-MIR393: a salinity- and alkaline stress-related microRNA gene. Mol Biol Rep. 2011;38:237–42.
Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–500.
Gautrat P, Laffont C, Frugier F. Compact root architecture 2 promotes root competence for nodulation through the miR2111 systemic effector. Curr Biol. 2020;30:1339–45.
Golicz AA, Singh MB, Bhalla PL. The long intergenic noncoding RNA (LincRNA) landscape of the soybean genome. Plant Physiol. 2018;176:2133–47.
Guan Q, Lu X, Zeng H, Zhang Y, Zhu J. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013;74:840–51. https://doi.org/10.1111/tpj.12169.
Guo J, Ren Y, Tang Z, Shi W, Zhou M. Characterization and expression profiling of the ICE-CBF-COR genes in wheat. PeerJ. 2019;7:81–90.
Gupta M, Das NU. Divergence in patterns of leaf growth polarity is associated with the expression divergence of miR396. Plant Cell. 2015;27:2785–99.
Hajyzadeh M, Turktas M, Khawar KM, Unver T. MiR408 overexpression causes increased drought tolerance in chickpea. Gene. 2015;555:186–93. https://doi.org/10.1016/j.gene.2014.11.002.
Hamilton AJ, Baulcombe DC. A novel species of small anti- sense RNA in post transcriptional gene silencing. Science. 1999;286:950–2. https://doi.org/10.1126/science.286.5441.950.
Hammond SM, Bernstein E, Beach D, Hannon GJ. AnRNA- directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–6. https://doi.org/10.1038/35005107.
Hamza NB, Sharma N, Tripathi A, Sanan-Mishra N. MicroRNA expression profiles in response to drought stress in Sorghum bicolor. Gene Expr Patterns. 2016;20(2):88–98.
Hatfield JL, Antle J, Garrett KA, Izaurralde RC, Mader T, Marshall E, Nearing M, Philip Robertson G, Ziska L. Indicators of climate change in agricultural systems. Clim Chang. 2018. https://doi.org/10.1007/s10584-018-2222-2.
Hibara K, et al. Arabidopsis CUP-SHAPED COTYLE-DON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell. 2006;18:2946–57.
Horiguchi G, et al. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 2005;43(68–78):57.
Huang L, Dong H, Zhou D, Li M, Liu Y, Zhang F, Feng Y, Yu D, Li S, Cao J. Syatematic identification of long non-coding RNAs during pollen development and fertilization in Brassica rapa. Plant J. 2018;96:203–22.
Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicerin the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. https://doi.org/10.1126/science.1062961.
Irish VF, Sussex IM. Function of the APETALA-1 gene during Arabidopsis floral development. Plant Cell. 1990;2:741–53.
Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Ann Rev Plant Biol. 2006;57:19–53. https://doi.org/10.1146/annurev.arplant.57.032905.105218.
Juarez MT, et al. MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–8.
Jung JH, et al. The miR172 target TOE3 represses AGAMOUS expression during Arabidopsis floral patterning. Plant Sci. 2014;215:29–38.
Kamthan A, Chaudhuri A, Kamthan M, Datta A. Small RNAs in plants: recent development and application for crop improvement. Front Plant Sci. 2015;6:208. https://doi.org/10.3389/fpls.2015.00208.
Khraiwesh B, Zhu JK, Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta. 2012;1819:137–48.
Kidner CA, Martienssen RA. Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature. 2004;428:81–4.
Kim JH, et al. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 2003;36:94–104.
Knauer S, et al. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meri-stem. Dev Cell. 2013;24:125–32.
Koyama T, et al. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell. 2010;22:3574–88.
Kumar R. Role of microRNAs in biotic and abiotic stress responses in crop plants. Appl Biochem Biotechnol. 2014;174:93–115.
Lämke J, Brzezinka K, Altmann S, Bäurle I. A hit-and-run heat shock factor governs sustained histone methylation and transcriptional stress memory. EMBO J. 2016;35:162–75. https://doi.org/10.15252/embj.201592593.
Lee H, Yoo SJ, Lee JH, Kim W, Yoo SK, Fitzgerald H, et al. Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. 2010;38:3081–93. https://doi.org/10.1093/nar/gkp1240.
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic genelin-encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. https://doi.org/10.1016/0092-8674(93)90529-Y.
Lee YS, et al. Rice miR172 induces flowering by suppressing OsIDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice. 2014;7:31.
Lewsey MG, Hardcastle TJ, Melnyk CW, Molnar A, Valli A, Urich MA, Nery JR, Baulcombe DC, Ecker JR. Mobile small RNAs regulate genome-wide DNA methylation. Proc Natl Acad Sci U S A. 2016;113:E801–10.
Li DD, Qiao HL, Qiu WJ, Xu X, Liu TM, Jiang QL, et al. Identification and functional characterization of intermediate-size non-coding RNAs in maize. BMC Genom. 2018;19:1–11.
Li J, Duan YJ, Sun NL, Wang L, Feng SS, Fang YJ, Wang YP. The miR169n-NF-YA8 regulation module involved in drought resistance in Brassica napus L. Plant Sci. 2021;313:111062.
Li S, Castillo-Gonzalez C, Yu B, Zhang X. The functions of plant small RNAs in development and in stress responses. Plant J. 2017;90:654–70.
Li W, Wang T, Zhang Y, Li Y. Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. J Exp Bot. 2016;67:175–94. https://doi.org/10.1093/jxb/erv450.
Li WX, Oono Y, Zhu JH, He XJ, Wu JM, Iida K, Lu XY, Cui XP, Jin HL, Zhu JK. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and post transcriptionally to promote drought resistance. Plant Cell. 2008;20:2238–51.
Li W, Cui X, Meng Z, Huang X, Xie Q, Wu H, et al. Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 2012;158:1279–92. https://doi.org/10.1104/pp.111.188789.
Li X, et al. Control of tillering in rice. Nature. 2003;422:618–21.
Lin JS, Kuo CC, et al. MicroRNA160 modulates plant development and heat shock protein gene expression to mediate heat tolerance in Arabidopsis. Front Plant Sci. 2018;9:68.
Liu DG, Mewalal R, Hu RB, Tuskan GA, Yang XH. New technologies accelerate the exploration of non-coding RNAs in horticultural plants. Hortic Res. 2017;4:17031.
Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, et al. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–45.
Liu H, et al. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 2014;165:160–74.
Liu Q, Yan S, Yang T, Zhang S, Chen Y, Liu B. Small RNAs in regulating temperature stress response in plants. J Integr Plant Biol. 2017;59:774–91. https://doi.org/10.1111/jipb.12571.
Liu Q, Yang T, Yu T, Zhang S, Mao X, Zhao J, Wang X, Dong J, Liu B. Integrating small RNA sequencing with qtl mapping for identification of miRNAs and their target genes associated with heat tolerance at the flowering stage in rice. Front Plant Sci. 2017;8:43.
Liu X, Zhang X, Sun B, Hao L, Liu C, Zhang D, Tang H, Li C, Li Y, Shi Y, et al. Genome-wide identification and comparative analysis of drought-related microRNAs in two maize inbred lines with contrasting drought tolerance by deep sequencing. PLoS ONE. 2019;14:e0219176.
Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–6. https://doi.org/10.1126/science.1076311.
Luan M, Xu M, Lu Y, Zhang Q, Zhang L, Zhang C, Fan Y, Lang Z, Wang L. Family-wide survey of miR169s and NF-Yas and their expression profiles response to abiotic stress in maize roots. PLoS ONE. 2014;9:91369.
Ma C, Burd S, Lers A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015;84:169–87.
Ma J, Zhao P, Liu S, Yang Q, Guo H. The control of developmental phase transitions by microRNAs and their targets in seed plants. Int J Mol Sci. 2020;21:1971. https://doi.org/10.3390/ijms21061971.
Ma X, Zhao F, Zhou B. The characters of non-coding RNAs and their biological roles in plant development and abiotic stress response. Int J Mol Sci. 2022;23:4124. https://doi.org/10.3390/ijms23084124.
Marin E, et al. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregu-latory network quantitatively regulating lateral root growth. Plant Cell. 2010;22:1104–17.
Martin A, et al. Graft-transmissible induction of potato tuberization by the microRNA miR172. Development. 2009;136:2873–81.
Maizel A, Markmann K, Timmermans M, Wachter A. To move or not to move: roles and specificity of plant RNA mobility. Curr Opin Plant Biol. 2020;57:52–60.
McConnell JR, Barton MK. Leaf polarity and meri- stem formation in Arabidopsis. Development. 1998;125:2935–42.
Melnyk CW, Molnar A, Baulcombe DC. Intercellular and systemic movement of RNA silencing signals. EMBO J. 2011;30:3553–63.
Meng XX, Li AX, Yu B, Li SJ. Interplay between miRNAs and lncRNAs: mode of action and biological roles in plant development and stress adaptation. Comput Struct Biotechnol. 2021;19:2567–74.
Mette MF, Vander Winden J, Matzke M, Matzke AJ. Short RNAs can identify new candidate transposable element families in Arabidopsis. Plant Physiol. 2002;130:6–9. https://doi.org/10.1104/pp.007047.
Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–5.
Montgomery TA, et al. Specificity of ARGONAUTE7– miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–41.
Nair SK, et al. Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleav-age. Proc Natl Acad Sci U S A. 2010;107:490–5.
Nelson JM, et al. Expression of a mutant maize gene in the ventral leaf epidermis is sufficient to signal a switch of the leaf’s dorsoventral axis. Development. 2002;129:4581–9.
Nikovics K, et al. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell. 2006;18:2929–45.
Ochando I, et al. Alteration of the shoot radial pattern in Arabidopsis thaliana by a gain-of-function allele of the class III HD-Zip gene INCURVATA4. Int J Dev Biol. 2008;52:953–61.
Ohama N, Sato H, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017;22:53–65. https://doi.org/10.1016/j.tplants.2016.08.015.
Ori N, et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet. 2007;39:787–91.
Pan WJ, Tao JJ, Cheng T, Bian XH, Wei W, Zhang WK, Ma B, Chen SY, Zhang JS. Soybean miR172a improves salt tolerance and can function as a long-distance signal. Mol Plant. 2016;9:1337–40.
Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, et al. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 2010;62:960–76. https://doi.org/10.1111/j.0960-7412.2010.04208.x.
Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, et al. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 2003;132:1382–90. https://doi.org/10.1104/pp.103.021980.
Park W, Li J, Song R, Messing J, Chen X. Carpelfactory, a dicer homolog, and HEN1, a novel protein, actin microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–95. https://doi.org/10.1016/S0960-9822(02)01017-5.
Parry G, et al. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci U S A. 2009;106:22540–5.
Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–49.
Peláez P, Trejo MS, Iñiguez LP, Estrada-Navarrete G, Covarrubias AA, Reyes JL, et al. Identification and characterization of microRNAs in Phaseolus vulgaris by high-throughput sequencing. BMC Genom. 2012;13:83. https://doi.org/10.1186/1471-2164-13-83.
Peng Y, Zhang X, Liu Y, Chen X. Exploring heat-response mechanisms of microRNAs based on microarray data of rice post-meiosis panicle. Int J Genomics. 2020;17:7582612. https://doi.org/10.1155/2020/7582612.
Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–79.
Peschansky VJ, Wahlestedt CWC. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9:3–12.
Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–65.
Rai MI, Alam M, Lightfoot DA, Gurha P, Afzal AJ. Classification and experimental identification of plant long non-coding RNAs. Genomics. 2019;111:997–1005.
Reyes JL, Chua NH. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007;49:592–606.
Rodriguez RE, et al. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010;137:103–12.
Si-Ammour A, Windels D, Arn-Bouldoires E, Kutter C, Ailhas J, Meins F Jr, Vazquez F. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 2011;157:683–91.
Shen X, He J, Ping Y, Guo J, Hou N, Cao F, Li X, Geng D, Wang S, Chen P, et al. The positive feedback regulatory loop of miR160-Auxin Response Factor 17- HYPONASTIC LEAVES 1 mediates drought tolerance in apple trees. Plant Physiol. 2021;188:1686–708.
Song JB, et al. Regulation of leaf morphology by micro- RNA394 and its target leaf curling responsiveness. Plant Cell Physiol. 2012;53:1283–94.
Song JB, Gao S, Wang Y, Li BW, Zhang YL, Yang ZM. miR394 and its target gene LCR are involved in cold stress response in Arabidopsis. Plant Gene. 2016;5:56–64.
Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Bäurle I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell. 2014;26:1792–807. https://doi.org/10.1105/tpc.114.123851.
Sun X, Zheng HX, Li JL, Liu LN, Zhang XS, Sui N. Comparative transcriptome analysis reveals new lncRNAs responding to salt stress in sweet sorghum. Front Bioeng Biotechnol. 2020;8:331.
Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001–19.
Swiezewski S, et al. Small RNA-mediated chromatin silencing directed to the 30 region of the Arabidopsis gene encoding the developmental regulator. FLC Proc Natl Acad Sci U S A. 2007;104:3633–8.
Tang Y, Du G, Xiang J, Hu C, Li X, Wang W, Zhu H, Qiao L, Zhao C, Wang J, et al. Genome-wide identification of auxin response factor (ARF) gene family and the miR160-ARF18-mediated response to salt stress in peanut (Arachis hypogaea L.). Genomics. 2022;114:171–84.
Tang Z, Zhang L, Xu C, Yuan S, Zhang F, Zheng Y, Zhao C. Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol. 2012;159:721–38.
Tilsner J, Nicolas W, Rosado A, Bayer EM. Staying tight: plasmodesmal membrane contact sites and the control of cell-to-cell connectivity in plants. Annu Rev Plant Biol. 2016;67:337–64.
Tsikou D, Yan Z, Holt DB, Abel NB, Reid DE, Madsen LH, Bhasin H, Sexauer M, Stougaard J, Markmann K. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science. 2018;362:233–6.
Waititu JK, Zhang C, Liu J, Wang H. Plant non-coding RNAs: origin, biogenesis, mode of action and their roles in abiotic stress. Int J Mol Sci. 2020;21:8401. https://doi.org/10.3390/ijms21218401.
Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–49.
Wang Q, et al. Divide et impera: boundaries shape the plant body and initiate new meristems. New Phytol. 2016;209:485–98.
Wang S, Sun X, Hoshino Y, Yu Y, Jia B, Sun Z, Sun M, Duan X, Zhu Y. MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in Rice (Oryza sativa L.). PLoSOne. 2014;9:e91357. https://doi.org/10.1371/journal.pone.0091357.
Wang Y, Gao L, Li J, Zhu B, Zhu H, Luo Y, et al. Analysis of long-non-coding RNAs associated with ethylene in tomato. Gene. 2018;674:151–60.
Wang W, Liu D, Chen DD, Cheng YY, Zhang XP, Song LR, Hu MJ, Dong J, Shen FF. MicroRNA414c affects salt tolerance of cotton by regulating reactive oxygen species metabolism under salinity stress. RNA Biol. 2019;16:362–75.
Wei W, Li G, Jiang X, Wang Y, Ma Z, Niu Z, et al. Small RNA and degradome profiling involved in seed development and oil synthesis of Brassica napus. PLoS ONE. 2018;13:e0204998.
Wollmann H, et al. On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development. 2010;137:3633–42.
Wu G, et al. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell. 2009;138:750–9.
Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–47.
Wu MF, et al. Arabidopsis microRNA167 controls pat-terns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133:4211–8.
Xia J, Zeng C, Chen Z, et al. Endogenous small-noncoding RNAs and their roles in chilling response and stress acclimation in Cassava. BMC Genomics. 2014;15:1–19.
Xia K, Wang R, Ou X, Fang Z, Tian C, Duan J, Wang Y, Zhang M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE. 2012;7:e30039.
Xia K, Zeng X, Jiao Z, Li M, Xu W, Nong Q, Mo H, Cheng T, Zhang M. Formation of protein disulfide bonds catalyzed by OsPDIL1;1 is mediated by MicroRNA5144-3p in rice. Plant Cell Physiol. 2018;59:331–42.
Xie K, et al. Genomic organization, differential expres-sion, and interaction of SQUAMOSA promoter-binding-like tran-scription factors and microRNA156 in rice. Plant Physiol. 2006;14:280–93.
Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE4functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:12984–9. https://doi.org/10.1073/pnas.0506426102.
Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, Sun Q. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 2010;10:1–11.
Yan K, Liu P, Wu CA, Yang GD, Xu R, Guo QH, Huang JG, Zheng CC. Stress-induced alternative splicing provides mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol Cell. 2012;48:521–31.
Yang C, Li D, Mao D, Liu X, Ji C, Li X, Zhao X, Cheng Z, Chen C, Zhu L. Overexpression of micro RNA 319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 2013;36:2207–18.
Yant L, et al. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell. 2010;22:2156–70.
Yu YH, Ni ZY, Wang Y, Wan HN, Hu Z, Jiang QY, Sun XJ, Zhang H. Overexpression of soybean miR169c confers increased drought stress sensitivity in transgenic Arabidopsis thaliana. Plant Sci. 2019;285:68–78.
Yuan J, Li J, Yang Y, Tan C, Zhu Y, Hu L, et al. Stress-responsive regulation of long non-coding RNA polyadenylation in Oryza sativa. Plant J. 2018;93:814–27.
Yuan S, Li Z, Li D, Yuan N, Hu Q, Luo H. Constitutive expression of rice microRNA528 alters plant development and enhances tolerance to salinity stress and nitrogen starvation in creeping bent grass. Plant Physiol. 2015;169:576–93.
Zou CL, Wang YB, Wang B, Liu D, Liu L, Gai ZJ, Li CF. Long non-coding RNAs in the alkaline stress response in sugar beet (Beta vulgaris L.). BMC Plant Biol. 2020;20:227.
Zhang F, Luo X, Zhou Y, Xie J. Genome-wide identification of conserved microRNA and their response to drought stress in Dongxiang wild rice (Oryza rufipogon Gri.). Biotechnol Lett. 2016;38:711–21.
Zhang J, Zhang H, Srivastava AK, Pan BJ, Fang J, Shi H, Zhu JK. Knockdown of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 2018;176:2082–94. https://doi.org/10.1104/pp.17.01432.
Zhang X, Wang W, Wang M, Zhan HY, Liu JH. The miR396b of Poncirus trifoliata functions in cold tolerance by regulating ACC oxidase gene expression and modulating ethylene–polyamine homeostasis. Plant Cell Physiol. 2016;57:1865–78. https://doi.org/10.1093/pcp/pcw108.
Zhang XP, Dong J, Deng FN, Wang W, Cheng YY, Song LR, Hu MJ, Shen J, Xu QJ, Shen FF. The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 2019;19:459.
Zhao J, He Q, Chen G, Wang L, Jin B. Regulation of non-coding RNAs in heat stress responses of plants. Front Plant Sci. 2016;7:1213. https://doi.org/10.3389/fpls.2016.01213.
Zhao J, Yuan S, Zhou YN, Li Z, Hu BFG, Liu H, Li S, Luo H. Transgenic creeping bent grass overexpressing Osa-miR393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnol J. 2018. https://doi.org/10.1111/pbi.12960.
Zhao L, et al. miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 2007;51:840–9.
Zhou M, Tang W. MicroRNA156 amplifies transcription factor-associated cold stress tolerance in plant cells. Mol Genet Genom. 2019;294:379–93.
Zhdanov VP. Stochastic bursts in the kinetics of gene expression with regulation by long non-coding RNAs. JETP Lett. 2010;92:410–5.
Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X. Intergenic transcription by RNA polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009;23:2850–60.
Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, Zhang X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145:242–56.
Zuo ZF, He W, Li J, Mo B, Liu L. Small RNAs: the essential regulators in plant thermotolerance. Front Plant Sci. 2021;12:726762. https://doi.org/10.3389/fpls.2021.726762.
Acknowledgements
The authors express their gratitude to the Council of Scientific & Industrial Research (CSIR)-University Grant Commission (UGC), New Delhi, India for financial support in the form of Senior Research Fellowship to Miss. Rinku Mondal (2018-MAY-351826) and Mr. Adwaita Das (09/025(0250)/2018-EMR-I)
The authors would like to acknowledge Department of Botany, The University of Burdwan for research and infrastructure facilities.
Author information
Authors and Affiliations
Contributions
RM and AD wrote the manuscript. AB provided the critical comments and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to disclose.
Additional information
Corresponding Editor: Sachin Rustagi; Reviewers: Anjana Rustagi, Neeraj Kumar Vasistha.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mondal, R., Das, A. & Bandyopadhyay, A. Implications of small RNAs in plant development, abiotic stress response and crop improvement in changing climate. Nucleus 66, 321–339 (2023). https://doi.org/10.1007/s13237-023-00447-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-023-00447-1