Abstract
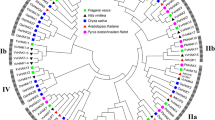
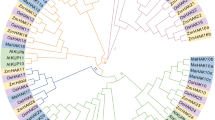
In plants, the HAK/KUP/KT family is the largest group of potassium transporters, and it plays an important role in mineral element absorption, plant growth, environmental stress adaptation, and symbiosis. Although these important genes have been investigated in many plant species, limited information is currently available on the HAK/KUP/KT genes for Pepper (Capsicum annuum L.). In the present study, a total of 20 CaHAK genes were identified from the pepper genome and the CaHAK genes were numbered 1 – 20 based on phylogenetic analysis. For the genes and their corresponding proteins, the physicochemical properties, phylogenetic relationship, chromosomal distribution, gene structure, conserved motifs, gene duplication events, and expression patterns were analyzed. Phylogenetic analysis divided CaHAK genes into four cluster (I–IV) based on their structural features and the topology of the phylogenetic tree. Purifying selection played a crucial role in the evolution of CaHAK genes, while whole-genome triplication contributed to the expansion of the CaHAK gene family. The expression patterns showed that CaHAK proteins exhibited functional divergence in terms of plant K+ uptake and salt stress response. In particular, transcript abundance of CaHAK3 and CaHAK7 was strongly and specifically up-regulated in pepper roots under low K+ or high salinity conditions, suggesting that these genes are candidates for high-affinity K+ uptake transporters and may play crucial roles in the maintenance of the Na+/K+ balance during salt stress in pepper. In summary, the results not only provided the important information on the characteristics and evolutionary relationships of CaHAKs, but also provided potential genes responding to potassium deficiency and salt stress.







Similar content being viewed by others
References
Ahn SJ, Shin R, Schachtman DP (2004) Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol 134:1135–1145. https://doi.org/10.1104/pp.103.034660
Amrutha RN, Sekhar PN, Varshney RK, Kishor PB (2007) Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci 172:708–721. https://doi.org/10.1016/j.plantsci.2006.11.019
Anschutz U, Becker D, Shabala S (2014) Going beyond nutrition: regulation of potassium homoeostasis as a common denominator of plant adaptive responses to environment. J Plant Physiol 171:670–687. https://doi.org/10.1016/j.jplph.2014.01.009
Aranda-Sicilia MN, Aboukila A, Armbruster U, Cagnac O, Schumann T, Kunz HH, Jahns P, Rodriguez-Rosales MP, Sze H, Venema K (2016) Envelope K+/H+ antiporters AtKEA1 and AtKEA2 function in plastid development. Plant Physiol 172:441–449. https://doi.org/10.1104/pp.16.00995
Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H (2012) ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res 40:W597-603. https://doi.org/10.1093/nar/gks400
Ashley MK, Grant M, Grabov A (2006) Plant responses to potassium deficiencies: a role for potassium transport proteins. J Exp Bot 57(2):425–436. https://doi.org/10.1093/jxb/erj034
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202-208. https://doi.org/10.1093/nar/gkp335
Cai K, Zeng F, Wang J, Zhang G (2021) Identification and characterization of HAK/KUP/KT potassium transporter gene family in barley and their expression under abiotic stress. BMC Genom 22:317. https://doi.org/10.1186/s12864-021-07633-y
Cellier F, Conejero G, Ricaud L, Luu DT, Lepetit M, Gosti F, Casse F (2004) Characterization of AtCHX17, a member of the cation/H+ exchangers, CHX family, from Arabidopsis thaliana suggests a role in K+ homeostasis. Plant J 39:834–846. https://doi.org/10.1111/j.1365-313X.2004.02177.x
Chen G, Hu Q, Luo L, Yang T, Zhang S, Hu Y, Yu L, Xu G (2015) Rice potassium transporter OsHAK1 is essential for maintaining potassium-mediated growth and functions in salt tolerance over low and high potassium concentration ranges. Plant Cell Environ 38:2747–2765. https://doi.org/10.1111/pce.12585
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Cheng X, Liu X, Mao W, Zhang X, Chen S, Zhan K, Bi H, Xu H (2018) Genome-Wide identification and analysis of HAK/KUP/KT potassium transporters gene family in wheat (Triticum aestivum L.). Int J Mol Sci. https://doi.org/10.3390/ijms19123969
Chérel I, Lefoulon C, Boeglin M, Sentenac H (2014) Molecular mechanisms involved in plant adaptation to low K(+) availability. J Exp Bot 65(3):833–848. https://doi.org/10.1093/jxb/ert402
Cuin TA, Miller AJ, Laurie SA, Leigh RA (2003) Potassium activities in cell compartments of salt-grown barley leaves. J Exp Bot 54:657–661. https://doi.org/10.1093/jxb/erg072
Desbrosses G, Kopka C, Ott T, Udvardi MK (2004) Lotus japonicus LjKUP is induced late during nodule development and encodes a potassium transporter of the plasma membrane. Mol Plant Microbe Interact 17:789–797. https://doi.org/10.1094/mpmi.2004.17.7.789
Edger PP, Pires JC (2009) Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res 17:699–717. https://doi.org/10.1007/s10577-009-9055-9
Edlund AF, Swanson R, Preuss D (2004) Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell 16(Suppl):S84-97. https://doi.org/10.1105/tpc.015800
Elumalai RP, Nagpal P, Reed JW (2002) A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 14:119–131. https://doi.org/10.1105/tpc.010322
Epstein E, Rains DW, Elzam OE (1963) Resolution of dual mechanisms of potassium absorption by barley roots. Proc Natl Acad Sci USA 5:684–692
Feng CZ, Luo YX, Wang PD, Gilliham M, Long Y (2021) MYB77 regulates high-affinity potassium uptake by promoting expression of HAK5. New Phytol. https://doi.org/10.1111/nph.17589
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M (2014) Pfam: the protein families database. Nucleic Acids Res 42:D222-230. https://doi.org/10.1093/nar/gkt1223
Genies L, Martin L, Kanno S, Chiarenza S, Carasco L, Camilleri V, Vavasseur A, Henner P, Leonhardt N (2021) Disruption of AtHAK/KT/KUP9 enhances plant cesium accumulation under low potassium supply. Physiol Plant 173(3):1230–1243
Gierth M, Maser P, Schroeder JI (2005) The potassium transporter AtHAK5 functions in K(+) deprivation-induced high-affinity K(+) uptake and AKT1 K(+) channel contribution to K(+) uptake kinetics in Arabidopsis roots. Plant Physiol 137:1105–1114. https://doi.org/10.1104/pp.104.057216
Grabov A (2007) Plant KT/KUP/HAK potassium transporters: single family - multiple functions. Ann Bot 99:1035–1041. https://doi.org/10.1093/aob/mcm066
Guether M, Balestrini R, Hannah M, He J, Udvardi MK, Bonfante P (2009) Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol 182:200–212. https://doi.org/10.1111/j.1469-8137.2008.02725.x
Gupta M, Qiu X, Wang L, Xie W, Zhang C, Xiong L, Lian X, Zhang Q (2008) KT/HAK/KUP potassium transporters gene family and their wholelife cycle expression profile in rice (Oryza sativa). Mol Genet Genom 280:437–452
Hajiahmadi Z, Abedi A, Wei H, Sun W, Ruan H, Zhuge Q, Movahedi A (2020) Identification, evolution, expression, and docking studies of fatty acid desaturase genes in wheat (Triticum aestivum L.). BMC Genom 21:778. https://doi.org/10.1186/s12864-020-07199-1
Han M, Wu W, Wu WH, Wang Y (2016) Potassium transporter KUP7 is involved in K+ acquisition and translocation in Arabidopsis root under K+-limited conditions. Mol Plant 9:437–446. https://doi.org/10.1016/j.molp.2016.01.012
He C, Cui K, Duan A, Zeng Y, Zhang J (2012) Genome-wide and molecular evolution analysis of the Poplar KT/HAK/KUP potassium transporter gene family. Ecol Evol 2:1996–2004. https://doi.org/10.1002/ece3.299
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Hyun TK, Rim Y, Kim E, Kim J-S (2014) Genome-wide and molecular evolution analyses of the KT/HAK/KUP family in tomato (Solanum lycopersicum L.). Genes Genom 36:365–374. https://doi.org/10.1007/s13258-014-0174-0
Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, dePamphilis CW (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–100. https://doi.org/10.1038/nature09916
Kaya C, Ashraf M, Alyemeni MN, Ahmad P (2020) The role of endogenous nitric oxide in salicylic acid-induced up-regulation of ascorbate-glutathione cycle involved in salinity tolerance of pepper (Capsicum annuum L.) plants. Plant Physiol Biochem 147:10–20. https://doi.org/10.1016/j.plaphy.2019.11.040
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lee YH, Kim HS, Kim JY, Jung M, Park YS, Lee JS, Choi SH, Her NH, Lee JH, Hyung NI, Lee CH, Yang SG, Harn CH (2004) A new selection method for pepper transformation: callus-mediated shoot formation. Plant Cell Rep 23:50–58. https://doi.org/10.1007/s00299-004-0791-1
Leigh RA, Wyn Jones RG (1984) A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol 97:1–13
Letunic I, Khedkar S, Bork P (2020) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. https://doi.org/10.1093/nar/gkaa937
Li XB, Fan XP, Wang XL, Cai. Yang WC. (2009) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17:859–875
Li J, Long Y, Qi GN, Li J, Xu ZJ, Wu WH, Wang Y (2014) The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex. Plant Cell 26:3387–3402. https://doi.org/10.1105/tpc.114.123455
Li W, Xu G, Alli A, Yu L (2018) Plant HAK/KUP/KT K(+) transporters: function and regulation. Semin Cell Dev Biol 74:133–141. https://doi.org/10.1016/j.semcdb.2017.07.009
Liu JL, Liu JJ, Chen AQ, Ji MJ, Chen JD, Yang XF, Gu M, Qu HY, Xu GH (2016a) Analysis of tomato plasma membrane H(+)-ATPase gene family suggests a mycorrhiza-mediated regulatory mechanism conserved in diverse plant species. Mycorrhiza 26(7):645–656. https://doi.org/10.1007/s00572-016-0700-9
Liu L, Zheng C, Kuang B, Wei L, Yan L, Wang T (2016b) Receptor-like kinase RUPO interacts with potassium transporters to regulate pollen tube growth and integrity in rice. PLoS Genet 12(7):e1006085. https://doi.org/10.1371/journal.pgen.1006085
Liu J, Liu J, Liu J, Cui M, Huang Y, Tian Y, Chen A, Xu G (2019) The potassium transporter SlHAK10 is involved in mycorrhizal potassium uptake. Plant Physiol 180:465–479. https://doi.org/10.1104/pp.18.01533
Lu Y, Chanroj S, Zulkifli L et al (2011) Pollen tubes lacking a pair of K+ transporters fail to target ovules in Arabidopsis. Plant Cell 23(1):81–93. https://doi.org/10.1105/tpc.110.080499
Luan S, Lan W, Chul LS (2009) Potassium nutrition, sodium toxicity, and calcium signaling: connections through the CBL-CIPK network. Curr Opin Plant Biol 12(3):339–346. https://doi.org/10.1016/j.pbi.2009.05.003
Luan M, Tang RJ, Tang Y, Tian W, Hou C, Zhao F, Lan W, Luan S (2017) Transport and homeostasis of potassium and phosphate: limiting factors for sustainable crop production. J Exp Bot 68(12):3091–3105. https://doi.org/10.1093/jxb/erw444
Ma X, Gai WX, Qiao YM, Ali M, Wei AM, Luo DX, Li QH, Gong ZH (2019) Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.). BMC Genom 20(1):775. https://doi.org/10.1186/s12864-019-6125-z
Maathuis FJ (2006) The role of monovalent cation transporters in plant responses to salinity. J Exp Bot 57:1137–1147. https://doi.org/10.1093/jxb/erj001
Maathuis FJ (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12:250–258. https://doi.org/10.1016/j.pbi.2009.04.003
Maher C, Stein L, Ware D (2006) Evolution of Arabidopsis microRNA families through duplication events. Genome Res 16:510–519. https://doi.org/10.1101/gr.4680506
Martínez-Cordero MA, Martínez V, Rubio F (2004) Cloning and functional characterization of the high-affinity K+ transporter HAK1 of pepper. Plant Mol Biol 56(3):413–421. https://doi.org/10.1007/s11103-004-3845-4
Martinez-Cordero MA, Martinez V, Rubio F (2005) High-affinity K+ uptake in pepper plants. J Exp Bot 56:1553–1562. https://doi.org/10.1093/jxb/eri150
Moscone EA, Scaldaferro MA, Grabiele M, Cecchini NM, Sánchez García Y, Jarret R, Daviña JR, Ducasse DA, Barboza GE, Ehrendorfer F (2007) The evolution of chili peppers (Capsicum Solanaceae): a cytogenetic perspectives, 745 edn. International Society for Horticultural Science (ISHS), Leuven
Mouline K, Véry AA, Gaymard F et al (2002) Pollen tube development and competitive ability are impaired by disruption of a Shaker K+ channel in Arabidopsis. Genes Dev 16(3):339–350. https://doi.org/10.1101/gad.213902
Nieves-Cordones M, Ródenas R, Chavanieu A, Rivero RM, Martinez V, Gaillard I, Rubio F (2016) Uneven HAK/KUP/KT protein diversity among angiosperms: species distribution and perspectives. Front Plant Sci 7:127
Okada T, Nakayama H, Shinmyo A, Yoshida K (2008) Expression of OsHAK genes encoding potassium ion transporters in rice. Plant Biotechnol 25:241–245. https://doi.org/10.5511/plantbiotechnology.25.241
Osakabe Y, Arinaga N, Umezawa T, Katsura S, Nagamachi K, Tanaka H, Ohiraki H, Yamada K, Seo SU, Abo M, Yoshimura E, Shinozaki K, Yamaguchi-Shinozaki K (2013) Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25:609–624. https://doi.org/10.1105/tpc.112.105700
Qiao X, Li M, Li L, Yin H, Wu J, Zhang S (2015) Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol 15:12. https://doi.org/10.1186/s12870-014-0401-5
Qin C, Yu C, Shen Y, Fang X, Chen L, Min J, Cheng J, Zhao S, Xu M, Luo Y, Yang Y, Wu Z, Mao L, Wu H, Ling-Hu C, Zhou H, Lin H, Gonzalez-Morales S, Trejo-Saavedra DL, Tian H, Tang X, Zhao M, Huang Z, Zhou A, Yao X, Cui J, Li W, Chen Z, Feng Y, Niu Y, Bi S, Yang X, Li W, Cai H, Luo X, Montes-Hernandez S, Leyva-Gonzalez MA, Xiong Z, He X, Bai L, Tan S, Tang X, Liu D, Liu J, Zhang S, Chen M, Zhang L, Zhang L, Zhang Y, Liao W, Zhang Y, Wang M, Lv X, Wen B, Liu H, Luan H, Zhang Y, Yang S, Wang X, Xu J, Li X, Li S, Wang J, Palloix A, Bosland PW, Li Y, Krogh A, Rivera-Bustamante RF, Herrera-Estrella L, Yin Y, Yu J, Hu K, Zhang Z (2014) Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl Acad Sci USA 111:5135–5140. https://doi.org/10.1073/pnas.1400975111
Rengel Z, Damon PM (2008) Crops and genotypes differ in efficiency of potassium uptake and use. Physiol Plant 133:624–636. https://doi.org/10.1111/j.1399-3054.2008.01079.x
Ruiz-Lau N, Bojorquez-Quintal E, Benito B, Echevarria-Machado I, Sanchez-Cach LA, Medina-Lara MF, Martinez-Estevez M (2016) Molecular cloning and functional analysis of a Na+-insensitive K+ transporter of Capsicum chinense Jacq. Front Plant Sci 7:1980. https://doi.org/10.3389/fpls.2016.01980
Saito T, Kobayashi NI, Tanoi K, Iwata N, Suzuki H, Iwata NTM (2013) Expression and functional analysis of the CorA-MRS2-ALR-type magnesium transporter family in rice. Plant Cell Physiol 54(10):1673–1683. https://doi.org/10.1093/pcp/pct112
Santa-María GE, Rubio F, Dubcovsky J, Rodríguez-Navarro A (1997) The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell 9:2281. https://doi.org/10.1105/tpc.9.12.2281
Schachtman DP, Schroeder JI (1994) Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370:655–658. https://doi.org/10.1038/370655a0
Shabala S, Cuin TA (2008) Potassium transport and plant salt tolerance. Physiol Plant 133:651–669. https://doi.org/10.1111/j.1399-3054.2007.01008.x
Shen Y, Shen L, Shen Z, Jing W, Ge H, Zhao J, Zhang W (2015) The potassium transporter OsHAK21 functions in the maintenance of ion homeostasis and tolerance to salt stress in rice. Plant Cell Environ 38:2766–2779. https://doi.org/10.1111/pce.12586
Song T, Shi Y, Shen L, Cao CJ, Shen Y, Jing W, Tian QX, Lin F, Li WY, Zhang WH (2021) An endoplasmic reticulum-localized cytochrome b 5 regulates high-affinity K(+) transport in response to salt stress in rice. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.2114347118
Templalexis D, Tsitsekian D, Liu C et al (2021) Potassium transporter TRH1/KUP4 contributes to distinct auxin-mediated root system architecture responses. Plant Physiol. https://doi.org/10.1093/phys/kiab472
Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI (2000) The Arabidopsis HKT1 gene homolog mediates inward Na(+) currents in xenopus laevis oocytes and Na(+) uptake in Saccharomyces cerevisiae. Plant Physiol 122:1249–1259. https://doi.org/10.1104/pp.122.4.1249
Wamser AF, Cecilio Filho AB, Nowaki RHD, Mendoza-Cortez JW, Urrestarazu M (2017) Influence of drainage and nutrient-solution nitrogen and potassium concentrations on the agronomic behavior of bell-pepper plants cultivated in a substrate. PLoS ONE 12:e0180529. https://doi.org/10.1371/journal.pone.0180529
Wang Y, Wu WH (2013) Potassium transport and signaling in higher plants. Annu Rev Plant Biol 64:451–476. https://doi.org/10.1146/annurev-arplant-050312-120153
Wang Y, Wu WH (2017) Regulation of potassium transport and signaling in plants. Curr Opin Plant Biol 39:123–128. https://doi.org/10.1016/j.pbi.2017.06.006
Wang Y, Lu J, Chen D, Zhang J, Qi K, Cheng R, Zhang H, Zhang S (2018) Genome-wide identification, evolution, and expression analysis of the KT/HAK/KUP family in pear. Genome 61:755–765. https://doi.org/10.1139/gen-2017-0254
White PJ (2013) Improving potassium acquisition and utilisation by crop plants. J Plant Nutr Soil Sci 176:305–316. https://doi.org/10.1002/jpln.201200121
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. https://doi.org/10.1093/molbev/msm088
Yang Z, Gao Q, Sun C, Li W, Gu S, Xu C (2009) Molecular evolution and functional divergence of HAK potassium transporter gene family in rice (Oryza sativa L.). J Genet Genomics 36:161–172. https://doi.org/10.1016/S1673-8527(08)60103-4
Yang T, Zhang S, Hu Y, Wu F, Hu Q, Chen G, Cai J, Wu T, Moran N, Yu L, Xu G (2014) The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol 166:945–959. https://doi.org/10.1104/pp.114.246520
Yang T, Feng H, Zhang S, Xiao H, Hu Q, Chen G, Xuan W, Moran N, Murphy A, Yu L, Xu G (2020a) The potassium transporter OsHAK5 alters rice architecture via ATP-dependent transmembrane auxin fluxes. Plant Commun 1:100052. https://doi.org/10.1016/j.xplc.2020.100052
Yang T, Lu X, Wang Y, Xie Y, Ma J, Cheng X, Xia E, Wan X, Zhang Z (2020b) HAK/KUP/KT family potassium transporter genes are involved in potassium deficiency and stress responses in tea plants (Camellia sinensis L.): expression and functional analysis. BMC Genom 21:556. https://doi.org/10.1186/s12864-020-06948-6
Zhang Z, Zhang J, Chen Y, Li R, Wang H, Wei J (2012) Genome-wide analysis and identification of HAK potassium transporter gene family in maize (Zea mays L.). Mol Biol Rep 39:8465–8473. https://doi.org/10.1007/s11033-012-1700-2
Zonia L, Munnik T (2007) Life under pressure: hydrostatic pressure in cell growth and function. Trends Plant Sci 12(3):90–97. https://doi.org/10.1016/j.tplants.2007.01.006
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (32002123), Special project on Science and Technology of Anhui Province, China (201903b06020017), Major Science and Technology Projects in Anhui Province (Grant No. 18030701214), and funded by the Biotechnology in Plant Protection of Ministry of Agriculture and Rural Affairs and Zhejiang Province.
Author information
Authors and Affiliations
Contributions
JRZ, GHQ, and JJL planned and designed the research; JRZ, JJL, CYL and JYL performed the experiments; XLL, JRZ and JZ analyzed the data; GHQ, and JJL wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, J., Qin, G., Liu, X. et al. Genome-wide identification and expression analysis of HAK/KUP/KT potassium transporter provides insights into genes involved in responding to potassium deficiency and salt stress in pepper (Capsicum annuum L.). 3 Biotech 12, 77 (2022). https://doi.org/10.1007/s13205-022-03136-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03136-z