Abstract
Background
Calcineurin B-like proteins (CBLs) are major Ca2+ sensors that interact with CBL-interacting protein kinases (CIPKs) to regulate growth and development in plants. The CBL-CIPK network is involved in stress response, yet little is understood on how CBL-CIPK function in pepper (Capsicum annuum L.), a staple vegetable crop that is threatened by biotic and abiotic stressors.
Results
In the present study, nine CaCBL and 26 CaCIPK genes were identified in pepper and the genes were named based on their chromosomal order. Phylogenetic and structural analysis revealed that CaCBL and CaCIPK genes clustered in four and five groups, respectively. Quantitative real-time PCR (qRT-PCR) assays showed that CaCBL and CaCIPK genes were constitutively expressed in different tissues, and their expression patterns were altered when the plant was exposed to Phytophthora capsici, salt and osmotic stress. CaCIPK1 expression changed in response to stress, including exposure to P. capsici, NaCl, mannitol, salicylic acid (SA), methyl jasmonate (MeJA), abscisic acid (ABA), ethylene (ETH), cold and heat stress. Knocking down CaCIPK1 expression increased the susceptibility of pepper to P. capsici, reduced root activity, and altered the expression of defense related genes. Transient overexpression of CaCIPK1 enhanced H2O2 accumulation, cell death, and expression of genes involved in defense.
Conclusions
Nine CaCBL and 26 CaCIPK genes were identified in the pepper genome, and the expression of most CaCBL and CaCIPK genes were altered when the plant was exposed to stress. In particular, we found that CaCIPK1 is mediates the pepper plant’s defense against P. capsici. These results provide the groundwork for further functional characterization of CaCBL and CaCIPK genes in pepper.
Similar content being viewed by others
Background
Plants have evolved tightly regulated signaling pathways to respond to the complex and varying environments that they are exposed to. Calcium (Ca2+) is a ubiquitous second messenger that regulates physiological and developmental processes in plants, and Ca2+ levels change when plants are exposed to biotic and abiotic stressors [1,2,3]. Ca2+ signatures, which are the distinct set of Ca2+ changes that occur in cells, are triggered by different stimuli, and the release of Ca2+ activates specific channels, pumps and transporters that are located at the various cellular membranes [4]. Changes in the intracellular concentration of Ca2+ is captured by Ca2+ sensors, which in turn regulate signaling pathways involved in plant growth and development, including calmodulins (CAMs), calcium-dependent protein kinases (CDPKs), and calcineurin B-like proteins (CBLs) [3].
CBLs have a domain called the EF-hand, which captures intracellular Ca2+. The EF-hand is a helix-loop-helix motif containing 12 residues of +X• + Y• + Z•-Y•-X••-Z, where the letters represent the ligands involved in metal coordination, and the dots represent the intervening residues [5]. CBL-interacting protein kinases (CIPKs) belong to the SnRK3 protein family that contain a Ser/Thr protein kinase domain [6]. CIPKs phosphorylate CBLs at a conserved Ser residue in CBL’s PFPF motif [7]. CIPKs frequently have a conserved N-terminal kinase domain, serine-threonine kinases domain, NAF/FISL motif and C-terminal regulatory domain [8, 9]. In addition to these domains, the C-terminus of CIPKs contain a protein-phosphatase interaction (PPI) domain, which interacts with phosphatase 2C (PP2C) proteins [10]. The interaction between the CBL and CIPK is involved in a Ca2+-decoding system called the CBL-CIPK network [11].
Previous studies have shown that the CBL-CIPK network is involved in regulating sodium (Na+), potassium (K+), magnesium (Mg2+) and nitrate (NO3−) transport across the plasma membrane (PM) or vacuolar membrane (tonoplast) [1, 12,13,14,15,16]. The CBL-CIPK network also plays an important role in regulating auxin, ABA signaling and stomatal movement [12, 17]. Abiotic stresses, such as ionic stress, osmotic stress and extreme temperatures, activate the salt overly sensitive (SOS) pathway and reactive oxygen species (ROS) signals to induce the expression of transcription factors involved in abiotic stress response [18]. The CBL-CIPK pathway was first identified in Arabidopsis through the discovery of the SOS pathway, which is comprised of AtCBL4 (SOS3), AtCIPK24 (SOS2) and the plasma membrane-localized Na+/H+ antiporter (SOS1) [19, 20]. AtCIPK24 interacts with CBLs to confer salt tolerance, and SOS1 is activated by CBL10-CIPK24 in a different salt-tolerance pathway on the tonoplast [21, 22]. The CBL1/CBL9-CIPK23 complexes are localized to the plasma membrane and regulate K+ in roots and stomatal guard cells by modulating the K+ channel Arabidopsis K+ Transporter1 (AKT1) [23, 24], and Arabidopsis plants overexpressing CIPK9 and CBL10 are sensitive to K+. In addition, CIPK9 interacts with CBL3 to regulate K+ homeostasis under low-K+ conditions [25, 26]. In Arabidopsis, the expression of CIPK3 changes in response to ABA, cold, and high salt stress conditions, and CIPK3 may be a cross-talk “node” that mediates interaction of ABA and abiotic stressors through ABA-dependent and ABA-independent pathways [27, 28]. Moreover, the CBL9-CIPK3-ABR1 (abscisic acid repressor 1) pathway regulates ABA and seed germination [29].
Recent studies have found that CIPKs regulate cell signaling across plants species. For instance, the apple CIPK protein kinase MdSOS2L1 interacts with MdCBL1, MdCBL4 and MdCBL10 to increase the levels of antioxidant metabolites and enhance salt tolerance in apple and tomato [30]. Overexpression of MdCIPK22 increases ABA sensitivity in an MdAREB2 (ABA responsive element binding factors) dependent manner, and MdCIPK13 phosphorylates MdSUT2.2 (sucrose transporter) to regulate salt tolerance [31, 32]. Overexpressing SlSOS2 (SlCIPK24) increases tolerance against salinity in tomato [33]. In wheat, TaCIPK23 is involved in ABA and drought stress responses, as well as the crosstalk between ABA signaling and drought [34].
Pepper (Capsicum annuum L.) is an important horticultural crop that belongs to Solanaceae, and is a rich source of vitamins, minerals and nutrients that are of great importance for human health [35, 36]. Nevertheless, the cultivation of pepper is threatened by biotic and abiotic stressors, such as pathogens, drought, salinity, and low temperature. Phytophthora capsici (P. capsici) is a devastating soil-borne pathogen that is causing significant damage to pepper crops worldwide by causing damping-off, seedling blight, and plant death [37, 38]. Infection of plants by pathogens leads to the activation of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI), and infection by microbes leads to micro-associated molecular patterns (MAMPs) of PTI [39]. OsCIPK14 and OsCIPK15 regulate MAMPs defense signaling pathways in rice, and TaCIPK5 positively regulates resistance against stripe rust fungus in wheat with TaCBL4 [40]. However, whether CBL and CIPK genes are involved in mediating stress response in pepper remains unclear, and studying the CBL-CIPK network is important for furthering our understanding of how pepper plants respond to biotic and abiotic stressors. Here, we identified two gene families (CaCBL and CaCIPK) in the pepper genome. Phylogenetic analyses were performed to investigate the evolutionary relationships of the nine CaCBL and 26 CaCIPK members. A comprehensive analysis of gene structure, protein motif conservation, chromosomal location, gene duplication, stress-related cis-elements, and prediction of protein-protein interaction networks were conducted to further understand the structure and relationship of CaCBL and CaCIPK genes. Furthermore, we examined the expression profiles of CaCBL and CaCIPK genes in pepper plants of various developmental stages and those exposed to biotic and abiotic stressors. We found that CaCIPK1 expression was induced by exposure to biotic and abiotic stressors, and we utilized the virus-induced gene silencing (VIGS) system to investigate the function of CaCIPK1 in pepper plants infected with P. capsici.
Results
Identification of CaCBL and CaCIPK genes in pepper
To identify CBLs and CIPKs in pepper, a HMM (Hidden Markov Model) analysis was performed against the CM334 (Capsicum annuum Cultivars in Mexico) and Zunla-1 (Capsicum annuum Cultivars in China) genomic databases. All putative genes were surveyed to verify the presence of conserved domains. Nine CBL and 26 CIPK genes were identified and named by chromosomal order (Additional file 1, Additional file 2). CaCBLs had four EF-hands, and CaCIPKs had a protein kinase catalytic domain (PKC), the 24-amino acid NAF/FISL motif and a PPI motif, similar to other genes in these families.
The length of CaCBL proteins varied from 205 (CaCBL3) to 258 (CaCBL5) amino acids, and the predicted molecular weights ranged from 23.55 to 29.73 kDa. The predicted isoelectric points (pI) were between 4.63 and 4.95. The length of CaCIPK proteins ranged from 335 (CaCIPK10) to 506 (CaCIPK26) amino acids, and the predicted molecular weights were between 38.27 and 57.26 kDa. The predicted instability indexes revealed some unstable proteins, inducing three CaCBLs (CaCBL5, − 7, − 9) and four CaCIPKs (CaCIPK5, − 10, − 15, − 26) (instability index =40). In the CaCBL family, two genes (CaCBL1 and CaCBL3) contained six introns, six genes (CaCBL2, − 4, − 6, − 7, − 8, − 9) had seven introns, and one gene (CaCBL5) had eight introns (Additional file 3A). In the CaCIPK family, seven CaCIPK genes (26.9%) (CaCIPK2, − 4, − 11, − 13, − 20, − 25, − 26) contained more than ten introns, while others had zero or one intron (Additional file 3B).
Phylogenetic and sequence analysis of CaCBL and CaCIPK genes
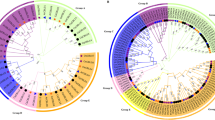
To understand the evolutionary relationship of CBL and CIPK proteins among pepper and other plants, a neighbor-joining (NJ) tree was constructed using amino acid sequences from Capsicum annuum, Arabidopsis thaliana, Manihot esculenta, Populus trichocarpa, Oryza sativa, Triticum aestivum, and Brassica napus (Fig. 1; Additional file 4). There were multiple CBL and CIPK sequences from wheat, potentially due to its hexaploidy, therefore only one sequence from each set of homologous gene sequences was used for the analysis. Sixty CBLs were clustered into four different groups (I to VI) with high bootstrap values (Fig. 1a). Three CBL genes (CaCBL3, CaCBL4 and CaCBL9) were categorized into group I, CaCBL5 was a part of group II, CaCBL1, − 2, and − 6 belonged to group III, and CaCBL7 and CaCBL8 clustered into group IV. CaCBL7 was orthologous to AtCBL7, and CaCBL8 was orthologous to wheat and rice CBLs. There were no sequences available for some CIPK genes, such as TaCIPK6, TaCIPK12, and TaCIPK13 [41].
The phylogenetic analysis of CBL (a) and CIPK (b) gene families from pepper, Arabidopsis cassava, canola, rice, poplar and wheat. Full-length protein sequences were used to construct Neighbor-Joining (NJ) trees by MEGA-X program with pairwise deletion, Poisson correction and bootstrap value 1000. Different species are denoted using different symbols. Subfamilies are distinguished with different colors. CBLs were classified into four groups, and CIPK were classified into five groups
A total 188 of CIPK genes from seven species were clustered into five subfamilies and assigned names (A to E) (Fig. 1b). Group A contained CaCIPK1, − 6, − 9, − 10, − 12, − 14, − 17, − 18, − 19, and − 21, group B included CaCIPK2, − 4, − 11, − 13, − 20, − 25, and − 26, group C included CaCIPK7 and CaCIPK8, group D contained only CaCIPK5, and group E contained CaCIPK3, − 15, − 16, − 22, − 23, and − 24. CaCIPK5 and CaCIPK20 were most similar to poplar and cassava CIPKs.
Multiple sequence alignment (MSA) revealed that CaCBLs have four EF-hand domains and a conserved PFPF motif (Additional file 5A). In addition, five CaCBLs harbored a conserved myristoylation motif (MGXXXS/T) on the N-terminus, including CaCBL1, − 2, − 3, − 4, and − 6. CaCBL5 had an extended N-terminus, which contained a transmembrane helix. CaCBL8 possessed the consensus motif for the tonoplast targeting sequences (TTS, MSQCXDGXKHXCXSXXXCF). MEME analysis showed that CaCBLs have six different conserved motifs, and these were named Motif-1 to Motif-6 (Fig. 2a; Additional file 6).
CaCIPKs contained a protein kinase domain, a unique NAF motif and PPI motif (Additional file 5B). MEME analysis revealed 10 conserved motifs for CaCIPKs, which were named motif-1 to motif-10 (Fig. 2b; Additional file 7). CaCIPK10 did not have motif-1 and motif-2, CaCIPK11 did not contain motif-3, CaCIPK23 did not contain motif-7 and motif-9, and CaCIPK2, − 4, − 7, − 8, − 9, − 10, − 24, and − 25 did not have motif-10. In addition, motif-8 was very similar to the NAF motif.
Chromosomal location and gene duplication of CaCBL and CaCIPK genes
CaCBL genes mapped onto six different chromosomes, including chromosome 1, 3, 6, 7, 10, and 12. Chromosome 1 contained three genes (CaCBL1, − 2, − 3), chromosome 3 had CaCBL4, chromosome 6 harbored CaCBL5 and CaCBL6, chromosome 7, − 10 and 12 contained CaCBL7, CaCBL8, CaCBL9, respectively (Fig. 3). CaCIPK genes were distributed across 12 chromosomes. Chromosome 1 and 2 both had three genes (CaCIPK1, − 2, − 3, and CaCIPK4, − 5, − 6), and chromosome 4, 6, and 9 had four genes each. Chromosome 5 contained two genes (CaCIPK12, − 13), and the other chromosomes had only one CaCIPKs.
Two segmental duplication events were predicted for CBLs (CaCBL1, − 2, − 6, and CaCBL7, − 8), and five duplication events were predicted for CIPKs (CaCIPK2, − 4, − 25, CaCIPK3, − 11, − 22, CaCIPK6, − 9, − 10, CaCIPK13, − 26, CaCIPK18, − 21) (Fig. 3). However, we did not identify any tandem duplication for CaCBL and CaCIPK genes. To explore the evolutionary constraints on CaCBL and CaCIPK gene families, the ratios of Ka (nonsynonymous) to Ks (synonymous substitutions) were estimated, and the results were less than 1 (Additional file 8). The chromosomal duplication events likely occurred between 106.34 (CaCIPK18/21) and 14.67 (CaCIPK9/10) million years ago.
Interaction network of CaCBL and CaCIPK members
To investigate the relationship of CaCBLs and CaCIPKs, a protein-protein interaction network was built using homologs of the Arabidopsis interaction network (Additional file 9). Nine CaCBLs were homologous to five AtCBLs, and 26 CaCIPKs were homologous to 13 AtCIPKs. The co-expression network of CBLs and CIPK was calculated using the Pearson correlation coefficient (PCC), where the red lines indicate PCC > 0, and the green lines indicate PCC < 0. The analysis inferred that CaCBL1, − 2, − 3, − 4, − 6 may interact with CaCIPK1, − 4, − 13, − 15, − 16, − 18, − 21, − 23, − 24, and − 26, and CaCBL7 and CaCBL8 may interact with CaCIPK1, − 2, − 3, − 4, − 5, − 7, − 8, − 13, − 15, − 16, − 18, − 21, − 22, − 23, − 24, − 25, and − 26. The analysis also predicted that CaCBL5 interacted with CaCIPK20, and CaCBL9 interacted with CaCIPK4 and CaCIPK11.
Cis-acting elements of CaCBL and CaCIPK genes in pepper
To better understand how CaCBL and CaCIPK genes are regulated, the 1500 bp upstream sequences of the coding region were detected by PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [42] to identify cis-acting elements (upstream sequences are shown in Additional file 10). Thirteen cis-elements, including ABRE (abscisic acid responsiveness), ARE (anaerobic induction), CE3 (ABA and VP1 responsiveness), CGTCA-motif (MeJA-responsiveness), GC-motif (enhancer-like element involved in anoxic specific inducibility), HSE (heat stress responsiveness), LTR (low-temperature responsiveness), MBS (MYB binding site involved in drought-inducibility), SARE and TCA-element (salicylic acid responsiveness), TC-rich (defense and stress), TGA-box (auxin-responsiveness) and WUN-motif (wound-responsiveness) were mapped onto the promoter regions (Fig. 4). ABRE elements were found in the promoter regions of five CaCBL genes (55.6%) including CaCBL1, − 3, − 7, − 8, and − 9, and 14 CaCIPK genes (53.8%) including CaCIPK1, − 2, − 3, − 5, − 6, − 11, − 15, − 16, − 18, − 20, − 21, − 22, − 23, and − 26. The TC-rich repeats elements were identified in the promoter regions of seven CaCBL (77.8%) and 21 CaCIPK genes (80.8%), excluding CaCBL7, − 8, CaCIPK1, − 12, − 14, − 16, and − 24. Interestingly, the promoter region of some CIPKs harbored at least one HSE elements, and the promoter of CaCIPK15 had the highest number of HSE elements (6). In addition, the WUN-motif was the least common element in these genes, and only in two CaCBL (CaCBL4, − 6) and five CaCIPK genes (CaCIPK3, − 9, − 12, − 18, − 22) contained WUN-motifs upstream of their promoter. All cis-elements that were identified in this analysis are involved in stress response and hormone signaling.
Predicted cis-acting regulatory elements in the promoter regions of CBL and CIPK genes. Promoter sequences (− 1500 bp) were analyzed by Plant CARE (Available online: http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Different shapes and colors represent different elements
Subcellular localization of CaCIPK proteins
To explore the potential function of CaCIPK, we selected one member from every subfamily to study its subcellular localization. The subcellular localizations were detected in epidermal cells of Nicotiana benthamiana by Agrobacterium-mediated transient expression. CaCIPK1 was localized to the nucleus, plasma membrane and cytoplasm, CaCIPK5 and CaCIPK20 were localized to the plasma membrane, and CaCIPK7 was localized to the nucleus and plasma membrane. In particular, the CaCIPK15-GFP fusion protein was localized to organelles or other structures in the cytoplasm (Additional file 11).
Gene expression patterns in response to various stresses and developmental stages of pepper
The CBL-CIPK network regulates stress response against biotic and abiotic stressors. In order to explore whether the expression of CaCBL and CaCIPK genes change when exposed to various stressors, including NaCl, mannitol, incompatible PC strain and compatible HX-9 strain of P. capsici, we conducted qRT-PCR analyses to study the expression patterns of CaCBLs and CaCIPKs. We chose samples at 6 h post treatment (hpt) for abiotic stress and 12 h post-inoculation (hpi) for biotic stress, and compared expression to plants that were collected at the same time that were not exposed to biotic or abiotic stress (Fig. 5a).
Expression patterns of CaCBLs and CaCIPKs in plants exposed to P. capsici, NaCl, and mannitol (a), and in different tissues (b). The grey means N/A. The samples in (a) were collected at different time points (6 hpt and 12 hpi for abiotic and biotic stresses respectively). “F” in (b) represents fruit. Actin (AY572427.1) is used as the internal control. Relative transcript levels were calculated using the comparative threshold (2−ΔΔCT) method, and normalized using log2. The heat map was created by MeV
Plants exposed to the incompatible PC strain of P. capsici showed up-regulation of six CaCBLs (CaCBL2, − 3, − 4, − 5, − 6, − 9) (log > 1) and down-regulation of one CaCBL (CaCBL8) (log < 1). CaCBL1 and CaCBL7 showed similar expression pattern as the control. When plants were exposed to the HX-9 strain, four CaCBLs (CaCBL2, − 3, − 4, − 5) were up-regulated and three (CaCBL6, − 7, − 9) were down-regulated. CaCBL1 and CaCBL8 showed similar expression pattern as the control. CaCBL3 and CaCBL4 were differentially expressed compared with CaCBL genes when plants exposed to P. capsici. CaCBL3 expression was strongly induced by exposure to PC (36.16-fold) and HX-9 (22.74-fold). In the CaCIPK family, 23 CaCIPKs were up-regulated when exposed to the PC strain. Differentially expressed genes included CaCIPK1, − 7, − 9, − 10, − 13, − 15, − 18, − 19, and − 20. CaCIPK1 expression was highly induced when exposed to the PC strain (51.68-fold) and HX-9 strain (16.34-fold). Eleven CaCIPKs were down-regulated when plants were treated with the HX-9 strain. CaCIPK8 was up-regulated when exposed to the PC strain and down-regulated when exposed to the HX-9 strain. Interestingly, CaCBL3/4 and CaCIPK1 were significantly up-regulated when plants were exposed to P. capsici. CaCBL9 and CaCIPK9 were up-regulated when exposed to the PC strain, but down-regulated when exposed to the HX-9 strain. Based the same expression patterns, we speculated that CaCBL3, − 4, and CaCIPK1, and CaCBL 9 and CaCIPK 9 were involved in the pepper’s resistance against P. capsici.
In response to abiotic stresses, five CaCBL genes were up-regulated when exposed to NaCl and seven CaCBL genes were up-regulated when exposed to mannitol. CaCBL3 was up-regulated by 4.25-fold when plants were exposed to NaCl and by 8.77-fold when exposed to mannitol. CaCBL2 and CaCBL9 were down-regulated when plants were treated with high levels of NaCl and mannitol. In the CaCIPK family, CaCIPK1, − 2, − 3, − 4, − 6, − 7, − 11, − 13, − 16, − 17, − 22, and − 25 showed differential expression levels when treated with NaCl or mannitol. For instance, CaCIPK1 was up-regulated by 2.13-fold when treated with NaCl and by 20.90-fold with mannitol. CaCIPK5, − 6, − 8, − 9, − 10, − 12, − 14, − 15, − 18, − 19, − 20, − 21, − 23, − 24, and − 26 were up-regulated in plants treated with mannitol (value ranged from 2.95 to 25.49-fold), but were not affected by NaCl stress. The same pattern was found in CaCBL8, indicating that they are co-regulated in response to osmotic stress.
To further investigate the spatial expression patterns of CaCBLs and CaCIPKs in pepper, qRT-PCR analysis was conducted on root, stem, leaf, flower, green fruit and red fruit. Expression levels of all genes were calculated relative to the expression levels of CaCIPK1 in the stem (Fig. 5b). CaCBL and CaCIPK genes were constitutively expressed in different tissues. Most CaCBL and CaCIPK genes showed higher expression in the root and leaf tissues compared with other tissues. For example, CaCBL4 expression was enriched in the root (95.80-fold) and leaf (24.98-fold), but was lower in stem (3.59-fold), green fruit (1.21-fold) and red fruit (1.19-fold). Expression levels of CaCBL1, − 5, − 7, and − 8, and CaCIPK17, and − 25 were high in all tissues. For instance, CaCIPK17 had the highest expression levels in all tissues compared to the other CaCBL and CaCIPK genes, especially in the root (662.41-fold). CaCIPK18 and CaCIPK25 were expressed in the red fruit (227.27-fold and 85.61-fold, respectively). These differential expression patterns of CaCBLs and CaCIPKs suggest that they may be involved in development and have tissue-specific functions.
Expression pattern of CaCIPK1 during stress and hormone treatment
Our results above showed that CaCIPK1 expression increased under different treatments, especially when plants were infected with P. capsici and exposed to high levels of mannitol (Fig. 5). To obtain better insight into the role of CaCIPK1 in mediating cell signaling and other stress response mechanisms, CaCIPK1 expression levels were measured in plants treated with SA, MeJA, ABA, ETH, and low and high temperatures. CaCIPK1 is a member of the CIPK family, which interacts with Ca2+ sensors. Therefore, leaves were sprayed with different concentrations of CaCl2 to explore whether CaCIPK1 was involved in the plant’s response to Ca2+ stress (Fig. 6).
Expression patterns of CaCIPK1 in plants exposed to SA, MeJA, ABA, ETH, cold (4 °C) and heat (42 °C) stress (a), and different concentration of CaCl2 (b). The samples in (a) were collected at different time points (0, 3, 6, 12, 24 hpt), and in (b) were at 6 hpt. Actin (AY572427.1) is used as the internal control. The error bars indicate the standard error (SE) for three replicates are showed. Letters (a–d) represent significant differences (LSD, p < 0.05)
CaCIPK1 expression was induced when leaves were sprayed with SA, MeJA, ABA and ETH. When sprayed with SA, CaCIPK1 was up-regulated and reached to a peak (6.36-fold) at 6 hpt, and was down-regulated at subsequent time points. When leaves were sprayed with ABA and ETH, the highest CaCIPK1 expression levels (3.46-fold and 3.77-fold respectively) were recorded at 6 hpt. Plants treated with MeJA showed highest CaCIPK1 expression level at 3 hpt (7.98-fold). In response to cold stress, CaCIPK1 expression steadily increased and reached highest expression level (3.28-fold) at 24 hpt. After heat stress, CaCIPK1 expression changed dynamically, and was significantly up-regulated (14.34-fold) at 24 hpt. In addition, expression of CaCIPK1 varied under different concentration of CaCl2 at 6 hpt. CaCIPK1 expression increased with exposure to higher concentrations of CaCl2, and reached a 5.55-fold up-regulation at 10 mM. CaCIPK1 expression was down-regulated when exposed to CaCl2 concentrations above 50 mM.
VIGS of CaCIPK1 increased pepper sensitivity to P. capsici
To further explore the function of CaCIPK1 in the pepper’s response to P. capsici infection, CaCIPK1 loss-of-function peppers were generated by VIGS using the AA3 cultivar. At 5 weeks post-inoculation, the positive control with pTRV2:CaPDS (phytoene desaturase gene) showed photobleaching phenotypes. qRT-PCR analysis confirmed that CaCIPK1 was silenced in the leaves, and CaCIPK1 expression in the silenced plants (pTRV2:CaCIPK1) was 50 to 80% lower than the negative control (pTRV2:00) (Fig. 7a). To ensure the construct specifically targeted CaCIPK1, the CaCIPK1 sequence was aligned with its homologous genes (CaCIPK17 and − 19) (Additional file 12A), and we found that the expression of CaCIPK17 and CaCIPK19 was not suppressed in pTRV2:CaCIPK1 plants (Additional file 12B). The sensitivity of CaCIPK1-silenced plants to the avirulent P. capsici (PC strain) was explored using detached leaves assays. Leaves from negative control and CaCIPK1-silenced plants were sampled to match the same region that showed photobleaching in the positive control sample. Leaves were injected with zoospore suspension of the PC strain. Slight disease symptoms were detected at 2-day post inoculation (dpi) in the CaCIPK1-silenced plants, and these symptoms gradually became apparent at 4 dpi. In contrast, very few disease lesions were observed in the control (Fig. 7b). Quantitative analysis of the lesion area revealed that CaCIPK1-silenced plants had significantly larger lesion areas (34.28%) than the control (1.42%) (Fig. 7c).
Knock-down of CaCIPK1 reduces pepper’s defense response to PC strain. (a) The relative expression of CaCIPK1, CaPR1, CaPO1, CaCAT, CaSAR8.2 and CaDEF1 in silenced and control plants; (b) Disease symptoms grow on the detached leaves of silenced and control plants at 4 day post inoculation (dpi). The scale bar represents 5 mm; (c) Percentage of the lesion area of silenced and control plants; (d) Root activity in CaCIPK1 silenced and control plants after inoculation with P. capsici. Error bars represent the mean ± SD of three independent biological replicates. Different letters (a–f) represent significant differences (LSD, p < 0.05)
To understand the molecular mechanisms underlying the increased sensitivity to the PC strain in the CaCIPK1 knocked down plants, expression levels of defense related genes were measured. Defense genes that were studied included CaPR1 (pathogenesis-related gene 1) [43], CaDEF1 (defensin gene) [44], CaSAR8.2 (systemic acquired resistance gene) [45], CaPO1 (peroxidase) [46], and CaCAT (catalase, highly homologous with AtCAT), and their expression patterns were examined at 2, 4, and 7 dpi (Fig. 7a). Knocking down CaCIPK1 resulted in a significant decline in the expression of CaPR1, CaSAR8.2 and CaDEF1. Control plants had 4.83-fold higher expression of CaPR1 than the CaCIPK1-silenced plants at 2 dpi. CaSAR8.2 and CaDEF1 showed 80% decrease in expression in CaCIPK1-silenced plants at both 2 dpi and 7 dpi. The expression of CaPO1 and CaCAT were significantly up-regulated in CaCIPK1-silenced plants, and the fold change was as high as 5-fold and 4-fold compared to the control at 4 dpi, respectively.
Furthermore, the vigor of the metabolism in the root system was measured by assessing root activity using triphenyltetrazolium chloride (TTC) [47]. TCC was reduced in the control and CaCIPK1-silenced plants that were treated with P. capsici (Fig. 7d). Root activity was reduced in both plants; however, CaCIPK1-silenced plants that were infected with P. capsici had significant lower root activity than control plants. The lowest activity was recorded in CaCIPK1-silenced plants at 4 dpi, where the activity was reduced by 57.3% compared to the control.
Transient expression of CaCIPK1 in pepper leaves
Knocking down CaCIPK1 increased the expression of CaPO1 and CaCAT, two genes that regulate ROS levels, therefore we explored whether CaCIPK1 is involved in ROS accumulation. The vectors 35S:00 and 35S:CIPK1 were ectopically overexpressed in pepper leaves using Agrobacterium. We measured cell death, H2O2 production, and expression of defense-related genes at 24 h post agroinfiltration. Transient expression of CaCIPK1 induced cell death and enhanced H2O2 accumulation compared with samples infected with the empty vector control (Fig. 8a, b). CaCIPK1 expression increased by 126.76-fold in the infected samples compared to the control, and CaPR1, CaDEF1, CaSAR8.2 expression increased by 16.11-fold, 3.96-fold and 136.54-fold, respectively (Fig. 8c).
Agrobacterium-mediated CaCIPK1 transient expression in pepper leaves. a Trypan blue staining of cell death in leaves; b DAB staining of H2O2 accumulation in leaves after agroinfiltration; (c) The relative expression of CaCIPK1, CaPR1, CaSAR8.2 and CaDEF1 in leaves. Data are means ± SD deviations from three independent experiments. Different letters indicate significant differences (LSD, P < 0.05). The scale bar represents 5 mm
Discussion
Calcium is a core regulator of plant development and responses to the environment [3]. The Ca2+ sensor CBLs and their target protein kinases CIPKs, comprise a complicated signaling network that allows plants to adapt to developmental and environmental stress. CBL-CIPK modules are involve in ion channel, phytohormones signaling network, growth and development [1, 12,13,14,15]. Past studies showed that multiple CBL and CIPK genes are found in many plant species, such as Arabidopsis thaliana (10 CBLs and 26 CIPKs), Manihot esculenta (8 CBLs and 26 CIPKs), Oryza sativa (10 CBLs and 34 CIPKs), Brassica napus (7 CBLs and 23 CIPKs), Populus trichocarpa (10 CBLs and 27 CIPKs), Triticum aestivum (24 CBLs and 79 CIPKs loci in sub-genomes A, B, and D respectively), and Vitis vinifera (8 CBLs and 20 CIPKs) [14, 40, 41, 48,49,50,51,52,53,54]. In this study, nine CBL and 26 CIPK genes were identified using the CM334 [55] and Zunla-1 [56] pepper genomes. MSA of CaCBLs found that all members contained four EF-hands, which consisted of 12 relatively conserved amino acids. Interestingly, the amino acids numbers between adjacent EF-hand were same. EF1 and EF2 were 23 amino acids apart, EF2 and EF3 were 25 amino acids apart, and 32 amino acids separated EF3 from EF4. The number of amino acids separating adjacent EF-hands were not consistent with findings from Arabidopsis thaliana, Oryza sativa and Vitis vinifera, where there were 22 amino acids between EF1 and EF2 [49, 52]. To further explore the numbers of amino acids between EF-hands in other species, CBLs in Arabidopsis thaliana, Capsicum annuum, Manihot esculenta, Populus trichocarpa, and Triticum aestivum were tested again for protein structure. We found there were consistently 23 amino acids between EF-1 and EF-2, unlike previous studies that identified 22 amino acids, and this is potentially due to differences in methods for counting amino acids between the domains (Additional file 13).
To uncover the potential function of CaCBLs and CaCIPKs, we performed phylogenetic analysis and MSA (Fig. 1; Additional file 5). Of the genes that belonged to group II (Fig. 1a), CaCBL5 had a transmembrane helix that may target the protein to the plasma membrane, similar to AtCBL10, OsCBL9, OsCBL10, and TaCBL9 [40]. Group IV contained the tonoplast targeting sequences (TTS). However, one member, CaCBL7, did not have the tonoplast targeting motif, similar to AtCBL7 [57, 58]. Compared with Arabidopsis, CaCBL8 and AtCBL2/3 contained TTS, suggesting that it is localized to the tonoplast [40, 59]. Additionally, five CaCBLs (CaCBL1, − 2, − 3, − 4 and − 6) had a myristoylation motif in the N-terminus, and this motif is required for binding to the membrane and interacting with SOS3 under salt stress [7, 60]. In addition, there was a PFPF motif at the C-terminus, and the motif contained a conserved serine residue known to interact with CIPKs. Interestingly, CIPKs in group B, such as CaCIPK2, − 4, − 11, − 13, − 20, − 25, and − 26, contained more than 10 introns, making this an intron-rich clade (Fig. 1b; Additional file 3). CaCIPKs in other groups contained zero or one intron, suggesting that they are an intron-poor clade. CIPK genes were also classified into intron-rich and intron-poor clades, such as in Arabidopsis thaliana, Triticum aestivum, and Oryza sativa [41, 52]. However, the number of introns in CBLs did not show a specific pattern. Hence the classification method using intron numbers is unique to the CIPK family. CaCIPKs had the conserved N-terminal kinase domain, C-terminal regulatory domain and NAF/FISL motif (Additional file 5B). The NAF domain consists of the conserved amino acids asparagine (N), alanine (A), and phenylalanine (F), and mediates the interaction with CBL. Next to the NAF motif, there was the PPI domain, which interacts with PP2C [8,9,10]. The N-terminal catalytic kinase domain has an ATP binding site and an activation loop [61].
In the gene duplication analysis, we found that the CaCBL family underwent two segmental duplication events (CaCBL1, − 2, − 6, and CaCBL7, − 8) in group III and group IV. The CaCIPK family harbored two segmental duplication events (CaCIPK6, − 9, − 10, and CaCIPK18, − 21) in group A, and three (CaCIPK2, − 4, − 25, CaCIPK3,-11,-22, and CaCIPK13, − 26) in group B. Tandem duplication events were not detected in CaCBL and CaCIPK genes. These results suggested that segmental duplications may have contributed to the complexity and diversity of both gene families. Additionally, the Ka/Ks ratio of CaCBL and CaCIPK genes (less than 1) inferred that the duplicated genes were maintained by purifying selection. The earliest duplication was for CaCIPK18 and CaCIPK21, and it was estimated to have occurred around 106.34 million years ago (Additional file 8).
The role of CBL-CIPK complexes in regulating plant development and response to environmental stress has been studied in different plant species, such as Arabidopsis and wheat [1, 3, 61]. To better understand the function of CaCBLs and CaCIPKs, we compared genes belonging to these families from pepper and Arabidopsis using a phylogenetic analysis (Additional file 14). CaCBL3 and CaCIPK20 were orthologous to AtCBL5 and AtCIPK24, respectively. AtCBL5-AtCIPK24 proteins are localized to the plasma membrane. Overexpression of AtCBL5 enhances salt and drought stress tolerance, and AtCIPK24 modulates cellular responses to salt stress and activate the Na+/H+ antiport activity of SOS1 [19, 21, 61, 62]. In pepper, CaCBL3 was up-regulated when exposed to salt (4.25-fold) and mannitol (8.77-fold) stress, and CaCIPK20 was also up-regulated when plants were exposed to mannitol (7.90-fold). MBS elements regulate gene expression under drought conditions, and these elements were found in the promoter region of CaCBL3 and CaCIPK20, suggesting that CaCBL3 may interact with CaCIPK20 to regulate drought stress tolerance (Figs. 4, 5a). PAT10 (Protein S-Acyl Transferase10)-CBL2/3-CIPK9/17 complexes regulate ABA signaling during stomatal movement in Arabidopsis [15, 17]. CaCBL7 and CaCBL8 were orthologous to AtCBL2 and AtCBL3, respectively. They contained ABRE elements in the promoter regions, suggesting that they may be regulated by PAT in ABA signaling. In wheat, TaCBL4 interacts with TaCIPK5 to positively modulate wheat resistance against fungus [40]. The genes orthologous to TaCBL4 and TaCIPK5, CaCBL9 and CaCIPK9/10, were up-regulated when plants were exposed to the PC strain, and down-regulated when exposed to the HX-9 strain. We speculate that CaCBL9-CaCIPK9/10 may regulate the pepper plant’s resistance against P. capsici.
The expression levels of CaCBLs and CaCIPKs were detected by qRT-PCR in plants exposed to pathogen, salt, and mannitol (Fig. 5a). However, expression patterns of CaCBL and CaCIPK genes were variable within the different subfamilies. For instance, CaCBL3 and CaCBL4 were up-regulated when exposed to stress, but CaCBL9 was up-regulated in plants treated with mannitol and incompatible P. capsici. CaCIPK13 was up-regulated by exposure to salt (10.87-fold), mannitol (89.38-fold) and incompatible P. capsici (35.20-fold), while CaCIPK26 was slightly up-regulated by exposure to mannitol (3.17-fold) and incompatible P. capsici (2.94-fold). CaCIPK9 was derived from a segmental duplication of CaCIPK10, and these two genes showed similar expression patterns. Nevertheless, other paralogs did not show similar patterns. In contrast, many CaCBLs and CaCIPKs genes were expressed at higher levels during different developmental stages (Fig. 5b). For instance, CaCIPK18 was expressed at the highest level in all stages, specifically in the red fruit (227.27-fold), inferring that it may have vital functions during different developmental stages. The expression level of CaCIPK4 was lower than other genes in all tissues, and its expression levels did not change when plants were exposed to P. capsici. Therefore, CBL-CIPK may play an important role in the response to biotic and abiotic stimuli, rather than development in pepper.
Five genes from each subfamily in CaCIPKs were chosen for subcellular localization assay. CaCIPK1 was localized to the nucleus, plasma membrane and cytoplasm, similar to TaCIPK14 and TaCIPK23 [34, 63]. Overexpression of TaCIPK14 enhances cold and salt stress tolerance in tobaccos. CaCIPK1 and TaCIPK14 belong to the same clade in Group I, therefore CaCIPK1 may also be involved in regulating tolerance against abiotic stress. CaCIPK5 and CaCIPK20 were localized to the plasma membrane. CaCIPK7 was localized to the nucleus and plasma membrane, similar to AtCIPK21, MeCIPK23 and BnaCIPK24 [64,65,66]. CaCIPK15 was targeted to organelles in the cytoplasm, deviating from the patterns observed in other CaCIPKs in this study (Additional file 11). In most plants, CIPKs are recruited by CBLs to the plasma membrane or tonoplatst to form a complex [21, 67], and it is likely that the interaction between CBLs and CIPKs influenced the plasma membrane localization of the five CIPKs were analyzed in this study.
While the role of the CBL-CIPK network in regulating response against abiotic stress is well documented, it remains unclear whether this network also regulates responses against biotic stress. OsCIPK14 and OsCIPK15 are rapidly induced by MAMPs in rice, and RNAi against these genes reduces sensitivity to Trichoderma viride/ethylene-inducing xylanase [68]. The TaCBL4-TaCIPK5 complex positively contributes to the interaction of wheat and Puccinia triiformis f. sp. tritici through ROS signaling [40]. CaCIPK1 was strongly up-regulation in the root of plants that were infected with incompatible (51.68-fold) and compatible (16.34-fold) P. capsici, indicating that CaCIPK1 may be involved in resistance against P. capsici in pepper. Moreover, the subcellular localization of CaCIPK1 was similar to TaCIPK14 and TaCIPK23. Overexpression of TaCIPK14 enhances cold and salt stress tolerance and TaCIPK23 positively regulates drought stress and ABA responses [34, 63], and CaCIPK1 may have similar functions. Meanwhile, the CaCIPK1 promoter region contained ABRE, HSE, MBS, LTR, and TCA cis-acting elements, which are involved in ABA signaling, heat stress, drought, low temperature and SA (Fig. 4). The presences of these elements suggest that CaCIPK1 is involved in abiotic and phytohormone stress response, and we tested the expression pattern of CaCIPK1 to verify this hypothesis (Fig. 6). SA, MeJA, ABA, and ETH are central regulators of defensive signaling and plant innate immunity [69, 70]. The results implied that CaCIPK1 may play a crucial role in defensive and innate immunity in pepper.
To confirm that CaCIPK1 is involved in mediating the interaction between pepper and incompatible P. capsici, TRV-VIGS was used to successfully knock down CaCIPK1. The silenced leaves were detached [71] and inoculated with the PC strain. There were larger lesions in the CaCIPK1-silenced plants compared to the control (24.06-fold), indicating that knocking down of CaCIPK1 increased sensitivity to incompatible P. capsici (Fig. 7c). Similar results were observed in wheat, where knockdown of TaCBL4 and TaCIPK5 reduced the defense response of wheat against stripe rust fungus [40]. To further study the role of CaCIPK1 in plants that were infected with the PC strain, root activity assays were conducted to detect the effects of the fungus on CaCIPK1-silenced plants. When the inoculation time was prolonged, root activity in CaCIPK1-silenced plants was lower than control plants, and there was a significant decrease in root activity at 4 and 7 dpi (Fig. 7b). These results showed that CaCIPK1-silenced plants had reduced resistance against incompatible P. capsici compared with control plants. In addition, CaPR1, CaSAR8.2, and CaDEF1 expression levels were lower in CaCIPK1-silenced plants that were exposed to PC (Fig. 7a). CaPR1 and CaSAR8.2 are genes involved in pathogenesis and systemic acquired resistance, and are induced by SA signaling when plants are exposed to biotic stress [40, 43]. Interestingly, CaCIPK1 expression was induced by SA (Fig. 6). Taken together, CaCIPK1 may be involved in the SA pathway to defend the pepper plant against incompatible P.capsici. CaCIPK1-silenced plants that were exposed to PC had higher expression levels of CaPO1 and CaCAT, which may lead to lower H2O2 accumulation. Previous studies have postulated that the accumulation of ROS (e.g., H2O2) and the release of Ca2+ positively regulate each other [72]. To verify whether CaCIPK1 is involved in H2O2 signaling, CaCIPK1 was transiently over-expressed in pepper. Plants over-expressing CaCIPK1 had higher cell death and H2O2 accumulation, and up-regulation of defense-related genes CaPR1 (16.11-fold) and CaSAR8.2 (136.54-fold) (Fig. 8). Thus, we hypothesize that CaCIPK1 is involved in H2O2 and SA signaling to modulate P. capsici tolerance by interacting with CBLs. Nevertheless, more studies are necessary to clarify the molecular mechanisms by which CaCIPK1 regulates the resistance of pepper against P. capsici.
Conclusions
The CBL-CIPK signaling pathway is a Ca2+-related pathway that regulates the plant’s response to environmental stimuli and ion stress [1]. Here, we identified nine CaCBL and 26 CaCIPK genes in pepper and most genes were highly expressed in different developmental stages. These genes also showed varying responses to biotic and abiotic stressors, suggesting that they may be crossing nodes of different signaling networks. Furthermore, CaCIPK1 expression levels changed in response to various stresses, including exposure to P. capsici, abiotic stress, and phytohormones. Knockdown of CaCIPK1 decreased the resistance of pepper against P. capsici, and changed the expression of defense related genes and root activity. Transient expression of CaCIPK1 in pepper leaves enhanced H2O2 accumulation and cell death. In brief, our study establishes a basic foundation for further research on the function of CaCBL and CaCIPK genes, and report a preliminarily exploration of the role of CaCIPK1 in pepper’s resistance to P. capsici. Further investigations are required to reveal the mechanism by which CaCIPK1 regulates resistance against P. capsici in pepper.
Methods
Genome-wide identification of CBL and CIPK genes in pepper
The proteome of pepper was downloaded from the Pepper Genome Database (CM334, http://peppergenome.snu.ac.kr/) and Zunla-1 (http://peppersequence.genomics.cn/). CBLs contain four unique EF-hands, and the CIPKs have a highly conserved protein kinase domain and NAF domain. The HMM profile of EF-hand (PF00036) and NAF (PF03822) were obtained from the Pfam (http://pfam.sanger.ac.uk/) protein family database. These domains were used as queries to search the pepper genome database with the BLASTP program (E-value ≤1.0E-3) [73]. All candidate CaCBLs were submitted to InterProScan (http://www.ebi.ac.uk/Tools/InterProScan) and SMART (http://smart.embl-heidelberg.de) to ensure the presence of the four EF-hand domains, and the same method was used to ensure CaCIPKs contained the Pkinase domain (IPR000719) and the NAF domain (IPR004041). Sequences were aligned with ClustalX (version 2.1) (http://www.clustal.org) to ensure that the CBL and CIPK genes that were identified in the analysis aligned to the CM334 and Zunla-1 genome sequences. When the sequences were different in CM334 and Zunla-1 databases, we designed the two sets of primers that were specific to the gene of interest in the different strains, and the PCR products were used to clone the gene. The sequences were aligned with ClustalX to identify the sequence of the gene of interest in cultivar AA3. The deduced amino acid and CDS sequences, theoretical isoelectric point (pI), instability index (with a value < 40 regarded as stable) [74] and protein molecular weight (MW) were analyzed by ExPASY (https://web.expasy.org/translate/; https://web.expasy.org/ protparam/). The WoLF PSORT program (http://wolfpsort.org/) was used to predict the subcellular localizations. Nomenclature of the CaCBL and CaCIPK genes were based on their chromosomal order.
Phylogenetic analysis
The full-length amino acid sequences of CBL and CIPK protein from Capsicum annuum, Arabidopsis thaliana, Manihot esculenta, Oryza sativa, Brassica napus, Populus trichocarpa, and Triticum aestivum, were aligned as an unrooted neighbor-joining phylogenetic tree using MEGA-X [75] with the bootstrap test replicated 1000 times. The classification of CaCBLs and CaCIPKs were based on previous research in Arabidopsis thaliana and Populus trichocarpa [50] and had high bootstrap values (> 50). The full-length amino acid sequences of genes from the species listed above were acquired from NCBI databases (http://www.ncbi.nlm.nih.gov/), Plant Genome Resource (https://phytozome.jgi.doe.gov/pz/portal.html), and TAIR database (https://www.arabidopsis.org/index.jsp). The sequences are displayed in Additional file 4.
Sequence analysis
The Multiple Sequence Alignment was executed using ClustalX to detect conserved domains. The position and number of introns in CaCBL and CaCIPK were visualized using Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/index.php) [76]. The conserved motifs of CaCBLs and CaCIPKs were identified and analyzed using protein sequences in the MEME online tool (http://meme-suite.org/tools/meme). The site distribution was selected as any number of repetitions, the optimum width of motifs ranged from 10 to 50 (10 to 200 for CIPK), and the maximum numbers of motifs were identified as 6 for CaCBLs and 10 for CaCIPKs. Motifs with position p-values less than 0.0001 are shown.
Chromosomal location and gene duplication
The chromosomal location of CaCBL and CaCIPK genes were identified using MapDraw [77]. Tandem duplications were defined as adjacent homologous genes on the same chromosome with a distance of less than 50-kb [78]. If they were paralogs located on duplicated chromosomal blocks, they were defined as a segmental duplication event [79]. The non-synonymous substitutions (Ka) and synonymous substitutions (Ks) were calculated by MEGA-X and DnaSP v6 [75, 80]. The divergence time (Mya, million years ago) was calculated as T = Ks / (2 × 6.1 × 10− 9) × 10− 6 [81].
Prediction of protein-protein interaction network
The protein-protein interaction relationships were tested to establish the genome-wide regulation network. Since there were no references about the interaction of CBL and CIPK proteins in pepper, homologous genes from Arabidopsis was used to predict the protein-protein interaction network for CBLs and CIPKs from pepper. First, CBL and CIPK proteins from Arabidopsis were analyzed using Arabidopsis Interactions Viewer (http://bar.utoronto.ca/interactions/cgi-bin/arabidopsis_interac tions_viewer.cgi). The homologs were identified in pepper, and the corresponding interaction network was created by Cytoscape 3.6.0 (National Institute of General Medical Sciences, MD, USA) [82].
Cis-acting elements in the promoters of CaCBL and CaCIPK genes
Regulatory elements of the promoter sequences can control gene expression [42]. The upstream regions (1500 bp) of CaCBL and CaCIPK genes were obtained from the PGD and Zunla-1 genomes, and regulatory elements, including ABRE (abscisic acid responsiveness), ARE (anaerobic induction), CE3 (ABA and VP1 responsiveness), CGTCA-motif (MeJA-responsiveness), GC-motif (enhancer-like element involved in anoxic specific inducibility), HSE (heat stress responsiveness), LTR (low-temperature responsiveness), MBS (MYB binding site involved in drought-inducibility), SARE and TCA-element (salicylic acid responsiveness), TC-rich (defense and stress), TGA-box (auxin-responsiveness) and WUN-motif (wound-responsiveness) were identified using the plant promoter database PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [42, 83].
Subcellular localization
The full-length cDNA sequences of CaCIPK1, CaCIPK5, CaCIPK7, CaCIPK15, and CaCIPK20 were cloned from the root tissue of AA3 and inserted into the pVBG2307 vector [84], which was modified with the green fluorescent protein (GFP) under the control of the 35S promoter. Constructs were introduced into the Agrobacterium strain GV3101. Agrobacterium cultures were grown overnight, and resuspended in 10 mM MES (pH 5.7) with 400 mM acetosyringone (3,5-dimethoxy-4′-hydroxy-acetophenone). Subcellular localization was observed following methods described by Wydro et al. [85]. The infiltrated Nicotiana benthamiana leaves were imaged 2 days after agroinfiltration using the OLYMPUS BX63 automated fluorescence microscope (OLYMPUS Corporation, Tokyo, Japan).
Plant materials, RNA extraction and quantitative RT-PCR
The pepper cultivar AA3 was obtained from the College of Horticulture, Northwest A&F University. AA3 is compatible with the HX-9 strain of P. capsici (virulent) and incompatible with the PC strain (avirulent). The plants were grown on soil and vermiculite (1:1), and cultivated in growth chambers (24/20 °C day/night temperature and 16/8 h day/night photoperiod). When plants reached the 6–8 true leaves stage, zoospore suspension of P. capsici was inoculated using the root drenching method as described previously [86]. Root samples were collected from treated (P. capsici) and control (treated with sterile water) plants, and collected at 0 and 12 hpi and immediately stored at − 80 °C. To induce NaCl and mannitol stress, seedlings at 6–8 true leaves stage were pre-hydroponically cultivated in sterile water for 2 days, and then treated with NaCl (200 mM) and mannitol (300 mM) hydroponically. Root samples were collected at 0 and 6 hpt [84]. Root, stem, leaf, flower, green fruits and red fruits were collected from normal AA3 plants grown in soil, frozen in liquid nitrogen and stored at − 80 °C for tissue-specific experiments.
Total RNA was extracted using the Trizol Reagent (Invitrogen, Carlsbad, CA), and reverse-transcribed using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Dalian, China). Primer Premier 5.0 was used to design primer pairs against CaCBL and CaCIPK genes for Quantitative Real-Time PCR analysis. The primer’s specificity was tested by NCBI Primer BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi) and checked by electrophoresis in 1.5% (w/v) agarose gel. All the primers used in this study are listed in Additional file 15. Actin was used as the reference gene [87]. Quantitative real-time PCR (qRT-PCR) was used to detect expression levels by SYBR Green Supermix (Takara, Dalian, China) on IQ5.0 Bio-Rad iCycler thermocycler (Bio-Rad, Hercules, CA, USA). All experiments included three independent biological replicates. The relative expression levels of pepper CBL and CIPK genes were calculated using the comparative 2−ΔΔCT method [88]. After normalizing dates by log2, a heatmap was drawn by Multi experiment viewer (MeV, http://www.tm4.org/mev.html).
Expression profile of CaCIPK1
The pepper line AA3 was grown to the 6–8 true leaf stage and sprayed with 5 mM SA, 50 μM MeJA, 0.57 μM ABA and 10 mM ETH solutions to detect changes in the expression level of CaCIPK1. Control plants were treated with sterile water. Plants that were at the same growth stage were placed at 4 °C and 42 °C for cold and heat stress, and leaves were collected at 0, 3, 6, 12, and 24 hpt. For CaCl2 stress, plants were sprayed with 0, 1, 10, 50, 100 mM CaCl2 and leaves were collected at 6 hpt. The samples were frozen in liquid nitrogen and stored at − 80 °C for RNA extraction.
Virus-induced gene silencing (VIGS) of CaCIPK1 in pepper
To construct the tobacco rattle virus (TRV) vector, primers for CaCIPK1 were designed by Sol Genomics Network (http://vigs.solgenomics.net/). A 245 bp fragment of the CaCIPK1 ORF was amplified using the specific primer pair vigs-F and vigs-R from cDNA isolated from the AA3 root grown under normal conditions (Additional file 12). The fragment was inserted into the original TRV vector for gene silencing. Agrobacterium strain GV3101 harboring pTRV1 was mixed at a 1:1 ratio with pTRV2 (negative control), pTRV2:CaPDS (silencing the phytoene desaturase gene, which induces bleaching) and pTRV2:CaCIPK1. The mixture was injected into fully extended cotyledons leaves [89]. Plants were cultivated in growth chambers (22/18 °C day/night temperature and 16/8 h day/night photoperiod). After 4 weeks, when the positive control (pTRV2:CaPDS) showed the photobleaching phenotype, leaf samples were collected from pTRV2:CaCIPK1 and pTRV2 to test the silencing efficiency by qRT-PCR. pTRV2:CaCIPK1 and control plants were treated with incompatible P. capsici (PC strain), and the third to fifth leaf from top of the control and CaCIPK1 silenced plants were picked. Leaves were washed with sterile water and injected with a 20 μL zoospore suspension (1 × 105 zoospores mL− 1) of incompatible P. capsici. Leaves were then moved into petri dishes and sealed with parafilm [86].
Agrobacterium-mediated transient expression assays
Agrobacterium strain GV3101 carrying 35S:00 (empty-vector) or 35S:CIPK1 was used for transient expression assays. Fresh pepper leaves were placed into resuspended 10 mM MES (pH 5.7, 200 mM acetosyringone) with 35S:00 and 35S:CIPK1, and vacuum infiltrated (OD600 = 1.0). Pepper leaves were photographed at 24 h after agroinfiltration using a Nikon D3300 (Nikon Corporation, Tokyo, Japan).
Histochemical analyses
Root activity was measured using TTC under a modified protocol by Wang et al. [90]. H2O2 production was detected by staining with 3,3′-diaminobenzidine (DAB). Cell death was visualized using trypan blue staining as described by Kim and Hwang [89].
Availability of data and materials
All the data in this research have been contained in the tables, figures, and files from the manuscript and additional files already. The sequences of Arabidopsis thaliana, Manihot esculenta, Populus trichocarpa, Oryza sativa, Triticum aestivum, and Brassica napus were acquired from National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/), the Plant Genome Resource (https://phytozome.jgi.doe.gov/pz/portal.html) and the TAIR database (https://www.arabidopsis.org/index.jsp).
Abbreviations
- ABA:
-
Abscisic acid
- ABR:
-
Abscisic acid repressor
- ABRE:
-
Abscisic acid responsiveness element
- AKT:
-
Arabidopsis K+ Transporter1
- AREB:
-
ABA resonsive element binding factor
- CAMs:
-
Calmodulins
- CBLs:
-
Calcineurin B-like proteins
- CDPKs:
-
Calcium-dependent protein kinases
- CIPKSs:
-
CBL-interacting protein kinases
- DAB:
-
3,3′-diaminobenzidine
- ETH:
-
Ethylene
- ETI:
-
Effector-triggered immunity
- GFP:
-
Green fluorescent protein
- H2O2 :
-
Hydrogen peroxide
- HMM:
-
Hidden Markov Model
- LTR:
-
Low-temperature responsiveness
- MAMPs:
-
Microbe-associated molecular patterns
- MBS:
-
MYB binding site involved in drought-inducibility
- MeJA:
-
Methyl Jasmonate
- MSA:
-
Multiple Sequence Alignment
- NJ tree:
-
Neighbor-Joining tree
- P. capsici :
-
Phytophthora capsici
- PAMPs:
-
Pathogen-associated molecular patterns
- PAT:
-
Protein S-acyl Transferase
- PCC:
-
Pearson Correlation Coefficient
- PDS:
-
Phytoene desaturase
- pI:
-
isoelectric points
- PKC:
-
Protein kinase catalytic domain
- PP2Cs:
-
Type 2C protein phosphatases
- PPI:
-
Protein-phosphatase interaction
- PTI:
-
Pattern-triggered immunity
- qRT-PCR:
-
Quantitative real-time PCR
- ROS:
-
Reactive oxygen species
- SA:
-
Salicylic acid
- SARE:
-
Salicylic acid responsiveness element
- SnRK3:
-
SNF1-related serine/threonine kinases
- SOS:
-
Salt overly sensitive
- SUT:
-
Sucrose transporter
- TRV:
-
Tobacco rattle virus
- TTC:
-
Triphenyltetrazolium Chloride
- TTS:
-
Tonoplast targeting sequences
- VIGS:
-
Virus-induced gene silencing
References
Luan S. The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 2009;14:37–42.
Kudla J, Becker D, Grill E, Hedrich R, Hippler M, Kummer U, et al. Advances and current challenges in calcium signaling. New Phytol. 2018;218:414–31.
Pandey GK, Kanwar P, Pandey A. Global comparative analysis of CBL–CIPK gene families in plants; 2014.
Dodd AN, Kudla J, Sanders D. The language of calcium signaling. Annu Rev Plant Biol. 2010;61:593–620.
Lewit-Bentley A, Réty S. EF-hand calcium-binding proteins. Curr Opin Struct Biol. 2000;10:637–43.
Shi J, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, et al. Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell. 1999;11:2393–405.
Du W, Lin H, Chen S, Wu Y, Zhang J, Fuglsang AT, et al. Phosphorylation of SOS3-like calcium-binding proteins by their interacting SOS2-like protein kinases is a common regulatory mechanism in Arabidopsis. Plant Physiol. 2011;156:2235–43.
Albrecht Â, Ritz O, Linder S, Harter K. The NAF domain defines a novel protein - protein interaction module conserved in Ca2+ −regulated kinases. EMBO J. 2001;20:1051–63.
Guo Y, Halfter U, Ishitani M, Zhu JK. Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell. 2001;13:1383–400.
Ohta M, Guo Y, Halfter U, Zhu J. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci. 2003;100:11771–6.
Luan S. Signalling drought in guard cells. Plant Cell Environ. 2002;25:229–37.
Weinl S, Kudla J. The CBL–CIPK Ca2+ −decoding signaling network : function and perspectives. New Phytol. 2009;184(3):517–28.
Thoday-Kennedy EL, Jacobs AK, Roy SJ. The role of the CBL-CIPK calcium signalling network in regulating ion transport in response to abiotic stress. Plant Growth Regul. 2015;76:3–12.
Zhang H, Yang B, Liu W-Z, Li H, Wang L, Wang B, et al. Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 2014;14:8.
Tang R-J, Zhao F-G, Garcia VJ, Kleist TJ, Yang L, Zhang H-X, et al. Tonoplast CBL–CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc Natl Acad Sci. 2015;112:3134–9.
Luan S, Lan W, Chul LS. Potassium nutrition, sodium toxicity, and calcium signaling: connections through the CBL-CIPK network. Curr Opin Plant Biol. 2009;12:339–46.
Song S-J, Feng Q-N, Li C, Li E, Liu Q, Kang H, et al. A tonoplast-associated calcium-signaling module dampens ABA signaling during stomatal movement. Plant Physiol. 2018;177:1666–78.
Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–24.
Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci. 2002;99:8436–41.
Liu J, Zhu J. A calcium sensor homolog required for plant salt tolerance. Science. 1998;280:1943–5.
Kim BG, Waadt R, Cheong YH, Pandey GK, Dominguez-Solis JR, Schültke S, et al. The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 2007;52:473–84.
Monihan SM, Magness CA, Yadegari R, Smith SE, Schumaker KS. Arabidopsis CALCINEURIN B-LIKE10 functions independently of the SOS pathway during reproductive development in saline conditions. Plant Physiol. 2016;171:369–79.
Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim BG, et al. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 2007;52:223–39.
Xue G, Lu LM, Yang TZ, Li XH, Xing XX, Xu SX. Enhanced tolerance to low-K+ stress in tobacco plants, that ectopically express the CBL-interacting protein kinase CIPK23 gene. Czech J Genet Plant Breed. 2016;52:77–82.
Liu L-L, Ren H, Chen L-Q, Wang Y, Wu W-H. A protein kinase, calcineurin B-like protein-interacting protein Kinase9, interacts with calcium sensor calcineurin B-like Protein3 and regulates potassium homeostasis under low-potassium stress in Arabidopsis. Plant Physiol. 2013;161:266–77.
Ren XL, Qi GN, Feng HQ, Zhao S, Zhao SS, Wang Y, et al. Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 2013;74:258–66.
Kim K-N, Cheong YH, Grant JJ, Pandey GK, Luan S. CIPK3 , a calcium sensor – associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell. 2003;15:411–23.
Pandey GK, Grant JJ, Cheong YH, Kim BG, Li LG, Luan S. Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol Plant. 2008;1:238–48.
Sanyal SK, Kanwar P, Yadav AK, Sharma C, Kumar A, Pandey GK. Arabidopsis CBL interacting protein kinase 3 interacts with ABR1, an APETALA2 domain transcription factor, to regulate ABA responses. Plant Sci. 2017;254:48–59.
Hu DG, Ma QJ, Sun CH, Sun MH, You CX, Hao YJ. Overexpression of MdSOS2L1, a CIPK protein kinase, increases the antioxidant metabolites to enhance salt tolerance in apple and tomato. Physiol Plant. 2016;156:201–14.
Ma QJ, Sun MH, Lu J, Liu YJ, You CX, Hao YJ. An apple CIPK protein kinase targets a novel residue of AREB transcription factor for ABA-dependent phosphorylation. Plant Cell Environ. 2017;40:2207–19.
Ma QJ, Sun MH, Kang H, Lu J, You CX, Hao YJ. A CIPK protein kinase targets sucrose transporter MdSUT2.2 at Ser254 for phosphorylation to enhance salt tolerance. Plant Cell Environ. 2018;42:1–13.
Egea I, Pineda B, Ortíz-Atienza A, Plasencia FA, Drevensek S, García-Sogo B, et al. The SlCBL10 calcineurin B-like protein ensures plant growth under salt stress by regulating Na+ and Ca2+ homeostasis. Plant Physiol. 2018;176:1676–93.
Cui XY, Du YT, J dong F, Yu TF, Wang CT, Chen M, et al. Wheat CBL-interacting protein kinase 23 positively regulates drought stress and ABA responses. BMC Plant Biol. 2018;18:1–13.
Lee YH, Kim HS, Kim JY, Jung M, Park YS, Lee JS, et al. A new selection method for pepper transformation: callus-mediated shoot formation. Plant Cell Rep. 2004;23:50–8.
Kim SH, Son GH, Bhattacharjee S, Kim HJ, Nam JC, Nguyen PDT, et al. The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 2014;78:978–89.
Liu P, Wei M, Zhang J, Wang R, Li B, Chen Q, et al. Changes in mycelia growth, sporulation, and virulence of Phytophthora capsici when challenged by heavy metals (Cu2+, Cr2+and Hg2+) under acid pH stress. Environ Pollut. 2018;235:372–80.
Lamour KH, Stam R, Jupe J, Huitema E, Programme PP, Road E, et al. The oomycete broad-host-range pathogen Phytophthora capsici. Mol Plant Pathol. 2012;13:329–37.
Boller T, Felix G. A Renaissance of Elicitors : Perception of Microbe-Associated Molecular Patterns and Danger Signals by Pattern-Recognition Receptors. Annu Rev Plant Biol. 2009;60:379–406.
Liu P, Duan Y, Liu C, Xue Q, Guo J, Qi T, et al. The calcium sensor TaCBL4 and its interacting protein TaCIPK5 are required for wheat resistance to stripe rust fungus. J Exp Bot. 2018;69:4443–57.
Sun T, Wang Y, Wang M, Li T, Zhou Y, Wang X, et al. Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 2015;15:269.
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–7.
Kim YJ, Hwang BK. Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol Plant. 2000;108:51–60.
Do HM, Lee SC, Jung HW, Sohn KH, Hwang BK. Differential expression and in situ localization of a pepper defensin (CADEF1) gene in response to pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Plant Sci. 2004;166:1297–305.
Lee SC, Hwang BK. Identification of the pepper SAR8.2 gene as a molecular marker for pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Planta. 2003;216:387–96.
Do HM, Hong JK, Jung HW, Kim SH, Ham JH, Hwang BK. Expression of peroxidase-like genes, H2O2 production, and peroxidase activity during the hypersensitive response to Xanthomonas campestris pv. Vesicatoria in Capsicum annuum. Mol Plant-Microbe Interact. 2003;16:196–205.
Comas LH, Eissenstat DM, Lakso AN. Assessing root death and root system dynamics in a study of grape canopy pruning. New Phytol. 2000;147:171–8.
Mo C, Wan S, Xia Y, Ren N, Zhou Y, Jiang X. Expression patterns and identified protein-protein interactions suggest that cassava CBL-CIPK signal networks function in responses to abiotic stresses. Front Plant Sci. 2018;9:1–17.
Xi Y, Liu J, Dong C, Cheng Z-M(M). The CBL and CIPK gene family in grapevine (Vitis vinifera): genome-wide analysis and expression profiles in response to various abiotic stresses. Front Plant Sci. 2017;8:1–15.
Yu Y, Xia X, Yin W, Zhang H. Comparative genomic analysis of CIPK gene family in Arabidopsis and Populus. Plant Growth Regul. 2007;52:101–10.
Hu W, Xia Z, Yan Y, Ding Z, Tie W, Wang L, et al. Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes. Front Plant Sci. 2015;6:914.
Kolukisaoglu Ü, Weinl S, Blazevic D, Batistic O, Kudla JJ, Kolukisaoglu U, et al. Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 2004;134:43–58.
Zhang H, Yin W, Xia X. Calcineurin B-like family in Populus: comparative genome analysis and expression pattern under cold, drought and salt stress treatment. Plant Growth Regul. 2008;56:129–40.
Xiang Y, Huang Y, Xiong L. Characterization of stress-responsive CIPK genes in Rice for stress tolerance improvement. Plant Physiol. 2007;144:1416–28.
Kim S, Park M, Yeom S-I, Kim Y-M, Lee JM, Lee H-A, et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet. 2014;46:270–8.
Qin C, Yu C, Shen Y, Fang X, Chen L, Min J, et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl Acad Sci. 2014;111:5135–40.
Tang RJ, Liu H, Yang Y, Yang L, Gao XS, Garcia VJ, et al. Tonoplast calcium sensors CBL2 and CBL3 control plant growth and ion homeostasis through regulating V-ATPase activity in Arabidopsis. Cell Res. 2012;22:1650–65.
Kleist TJ, Spencley AL, Luan S. Comparative phylogenomics of the CBL-CIPK calcium-decoding network in the moss Physcomitrella, Arabidopsis, and other green lineages. Front Plant Sci. 2014;5:1–17.
Batistič O, Rehers M, Akerman A, Schlücking K, Steinhorst L, Yalovsky S, et al. S-acylation-dependent association of the calcium sensor CBL2 with the vacuolar membrane is essential for proper abscisic acid responses. Cell Res. 2012;22:1155–68.
Saito S, Hamamoto S, Moriya K, Matsuura A, Sato Y, Muto J, et al. N -myristoylation and S -acylation are common modifications of Ca2+ −regulated Arabidopsis kinases and are required for activation of the SLAC1 anion channel. New Phytol. 2018;218:1504–21.
Sanyal SK, Pandey A, Pandey GK. The CBL-CIPK signaling module in plants: a mechanistic perspective. Physiol Plant. 2015;155:89–108.
Cheong YH, Sung SJ, Kim BG, Pandey GK, Cho JS, Kim KN, et al. Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in Arabidopsis. Mol Cells. 2010;29:159–65.
Deng X, Zhou S, Hu W, Feng J, Zhang F, Chen L, et al. Ectopic expression of wheat TaCIPK14, encoding a calcineurin B-like protein-interacting protein kinase, confers salinity and cold tolerance in tobacco. Physiol Plant. 2013;149:367–77.
Pandey GK, Kanwar P, Singh A, Steinhorst L, Pandey A, Yadav AK, et al. Calcineurin B-like protein-interacting protein kinase CIPK21 regulates osmotic and salt stress responses in Arabidopsis. Plant Physiol. 2015;169:780–92.
Yan Y, He X, Hu W, Liu G, Wang P, He C, et al. Functional analysis of MeCIPK23 and MeCBL1/9 in cassava defense response against Xanthomonas axonopodis pv. Manihotis. Plant Cell Rep. 2018;37:887–900.
Liu WZ, Deng M, Li L, Yang B, Li H, Deng H, et al. Rapeseed calcineurin B-like protein CBL4, interacting with CBL-interacting protein kinase CIPK24, modulates salt tolerance in plants. Biochem Biophys Res Commun. 2015;467:467–71.
Weinl S, Kudla J. The CBL-CIPK Ca2+-decoding signaling network : function and perspectives. New Phytol. 2009;184:517–28.
Kurusu T, Hamada J, Nokajima H, Kitagawa Y, Kiyoduka M, Takahashi A, et al. Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, Phytoalexin production, and defense gene expression by Calcineurin B-like protein-interacting protein kinases, OsCIPK14/15, in Rice cultured cells. Plant Physiol. 2010;153:678–92.
Lee SC, Luan S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012;35:53–60.
Rojo E, Solano R, Sánchez-Serrano JJ. Interactions between signaling compounds involved in plant defense. J Plant Growth Regul. 2003;22:82–98.
Bundó M, Coca M. Enhancing blast disease resistance by overexpression of the calcium-dependent protein kinase OsCPK4 in rice. Plant Biotechnol J. 2016;14:1357–67.
Steinhorst L, Kudla J. Calcium and reactive oxygen species rule the waves of signaling. Plant Physiol. 2013;163:471–85.
Guo M, Zhai Y, Lu J, Chai L, Chai W, Gong Z. Characterization of CaHsp70–1 , a Pepper Heat-Shock Protein Gene in Response to Heat Stress and Some Regulation Exogenous Substances in Capsicum annuum L. Int J Mol Sci. 2014;15:19741–59.
Guruprasad K, Reddy BVB, Pandit MW. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng Des Sel. 1990;4:155–61.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X : Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35:1547–9.
Guo M, Liu J-H, Lu J-P, Zhai Y-F, Wang H, Gong Z-H, et al. Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress. Front Plant Sci. 2015;6:806.
Liu RH, Meng JL. MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas. 2003;25:317–21.
Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;21:1–21.
Guo M, Liu JH, Ma X, Zhai YF, Gong ZH, Lu MH. Genome-wide analysis of the Hsp70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Sci. 2016;252:246–56.
Rozas J, Ferrer-mata A, JC S, Guirao-rico S, Librado P, Ramos-onsins E, et al. DnaSP 6 : DNA Sequence Polymorphism Analysis of Large Data Sets. Mol Biol Evol. 2017;34:3299–302.
Guo M, Lu J-P, Zhai Y-F, Chai W-G, Gong Z-H, Lu M-H. Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol. 2015;15:151.
Shannon P, Markiel A, Owen Ozier 2, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94.
Guo WL, Chen RG, Du XH, Zhang Z, Yin YX, Gong ZH, et al. Reduced tolerance to abiotic stress in transgenic Arabidopsis overexpressing a Capsicum annuum multiprotein bridging factor 1. BMC Plant Biol. 2014;14:1–13.
Wydro M, Kozubek E, Lehmann P. Optimization of transient agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim Pol. 2006;53:289–98.
Jin J-H, Zhang H-X, Tan J-Y, Yan M-J, Li D-W, Khan A, et al. A New Ethylene-Responsive Factor CaPTI1 Gene of Pepper (Capsicum annuum L.) Involved in the Regulation of Defense Response to Phytophthora capsici. Front Plant Sci. 2016;6:1–12.
Wan H, Yuan W, Ruan M, Ye Q, Wang R, Li Z, et al. Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem Biophys Res Commun. 2011;416:24–30.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–8.
Kim NH, Hwang BK. Pepper pathogenesis-related protein 4c is a plasma membrane-localized cysteine protease inhibitor that is required for plant cell death and defense signaling. Plant J. 2015;81:81–94.
Wang J-E, Li D-W, Zhang Y-L, Zhao Q, He Y-M, Gong Z-H. Defence responses of pepper (Capsicum annuum L.) infected with incompatible and compatible strains of Phytophthora capsici. Eur J Plant Pathol. 2013;136:625–38.
Acknowledgments
The authors thank to all members for assistance.
Funding
This work was supported through funding from National Key R&D Program of China (No. 2016YFD0101900), the National Natural Science Foundation of China (No. U1603102, No. 31772309), Independent Innovation Fund Project of Agricultural Science and Technology in Jiangsu [No.CX (17)3040].
Author information
Authors and Affiliations
Contributions
MX and ZHG conceived and designed the experiments. MX performed the experiments. WXG, YMQ and QHL analyzed the data. MX and MA performed in-silico analysis. MX wrote the paper. ZHG, AMW and DXL contributed reagents/ materials/ an analysis tools. MX and ZHG revised the paper. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
List of the identified CBL genes in pepper.
Additional file 2.
List of the identified CIPK genes in pepper.
Additional file 3.
Phylogenetic relationship and gene structure of CBL (A) and CIPK (B) in pepper.
Additional file 4.
Amino acid sequences of phylogenetic analysis.
Additional file 5.
The multiple sequence alignment of CaCBL and CaCIPK families.
Additional file 6.
MEME sequence of CaCBL.
Additional file 7.
MEME sequence of CaCIPK.
Additional file 8.
The Ka/Ks ratios and estimated divergence time for duplicated CaCBL and CaCIPK genes.
Additional file 9.
The predicative interaction network of CBL and CIPK genes in pepper according to the orthologs in Arabidopsis.
Additional file 10.
1500 bp promoter sequences of CaCBL and CaCIPK genes.
Additional file 11.
Subcellular localization of CaCIPK1, CaCIPK5, CaCIPK7, CaCIPK15 and CaCIPK20 in N. benthamiana epidermal cells.
Additional file 12.
The multiple sequence alignment of CaCIPK1, CaCIPK17, and CaCIPK19 (A) and expression patterns of CaCIPK17 and CaCIPK19 in CaCIPK1-silenced plants (B).
Additional file 13.
The multiple sequence alignment of CBLs from pepper, Arabidopsis, cassava, poplar and wheat.
Additional file 14.
The phylogenetic analysis of CBL (A) and CIPK (B) gene families from pepper and Arabidopsis.
Additional file 15.
Sequences of the primers used in this study.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Ma, X., Gai, WX., Qiao, YM. et al. Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.). BMC Genomics 20, 775 (2019). https://doi.org/10.1186/s12864-019-6125-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-6125-z