Abstract
Potassium is the most abundant inorganic cation that constitutes up to 10% of the total plant dry weight and plays a prominent role in plant growth and development. Plants exhibit a complex but highly organized system of channels and transporters, which are involved in absorption and distribution of K+ from soil to different parts of plants. In this study, we explored the K+ transport system in chickpea genome and identified 36 genes encoding potassium channels and transporters. The identified genes were further classified on the basis of their domain structure and conserved motifs. It includes K+ transporters (23 genes: 2 HKTs, 6 KEAs, and 15 KUP/HAK/KTs) and K+ channels (13 genes: 8 Shakers and 5 TPKs). Chromosomal localization of these genes demonstrated that various K+ transporters and channels are randomly distributed across all the eight chromosomes. Comparative phylogenetic analysis of K+ transport system genes from Arabidopsis thaliana, Glycine max, Medicago truncatula, and Oryza sativa revealed their strong conservation in different plant species. Similarly, gene structure analysis displayed conservation of family-specific intron/exon organization in the K+ transport system genes. Evolutionary analysis of these genes suggested the segmental duplication as principal route of expansion for this family in chickpea. Several abiotic stress-related cis-regulatory elements were also identified in promoter regions suggesting their role in abiotic stress tolerance. Expression analysis of selected genes under drought, heat, osmotic, and salt stress demonstrated their differential expression in response to these stresses. This signifies the importance of these genes in the modulation of stress response in chickpea. Present study provides the first insight into K+ transport system in chickpea and can serve as a basis for their functional analysis.







Similar content being viewed by others
References
Ahmad I, Mian A, Maathuis FJM (2016) Overexpression of the rice AKT1 potassium channel affects potassium nutrition and rice drought tolerance. J Exp Bot 67:2689–2698. https://doi.org/10.1093/jxb/erw103
Almeida P, Katschnig D, de Boer A (2013) HKT transporters—state of the art. Int J Mol Sci 14:20359–20385. https://doi.org/10.3390/ijms141020359
Amrutha RN, Sekhar PN, Varshney RK, Kishor PBK (2007) Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci 172:708–721. https://doi.org/10.1016/j.plantsci.2006.11.019
Assaha DVM, Ueda A, Saneoka H, al-Yahyai R, Yaish MW (2017) The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol 8. https://doi.org/10.3389/fphys.2017.00509
Bañuelos MA, Garciadeblas B, Cubero B, Rodríguez-Navarro A (2002) Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol 130:784–795. https://doi.org/10.1104/pp.007781
Becker D, Hoth S, Ache P, Wenkel S, Roelfsema MRG, Meyerhoff O, Hartung W, Hedrich R (2003) Regulation of the ABA-sensitive Arabidopsis potassium channel gene GORK in response to water stress. FEBS Lett 554:119–126. https://doi.org/10.1016/S0014-5793(03)01118-9
Beers MF, Zhao M, Tomer Y, Russo SJ, Zhang P, Gonzales LW, Guttentag SH, Mulugeta S (2013) Disruption of N-linked glycosylation promotes proteasomal degradation of the human ATP-binding cassette transporter ABCA3. Am J Physiol Cell Mol Physiol 305:L970–L980. https://doi.org/10.1152/ajplung.00184.2013
Clarkson DT, Hanson JB (1980) The mineral nutrition of higher plants. Annu Rev Plant Physiol Plant Mol Biol 31:239–298
Crooks GE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190. https://doi.org/10.1101/gr.849004
Cuéllar T, Pascaud F, Verdeil J-L et al (2010) A grapevine shaker inward K(+) channel activated by the calcineurin B-like calcium sensor 1-protein kinase CIPK23 network is expressed in grape berries under drought stress conditions. Plant J 61:58–69. https://doi.org/10.1111/j.1365-313X.2009.04029.x
Czempinski K, Gaedeke N, Zimmermann S, Müller-Röber B (1999) Molecular mechanisms and regulation of plant ion channels. J Exp Bot 50:955–966. https://doi.org/10.1093/jxb/50.Special_Issue.955
Dauterive R, Laroux S, Bunn RC, Chaisson A, Sanson T, Reed BC (1996) C-terminal mutations that alter the turnover number for 3-O-methylglucose transport by GLUT1 and GLUT4. J Biol Chem 271:11414–11421. https://doi.org/10.1074/jbc.271.19.11414
Davies C, Shin R, Liu W, Thomas MR, Schachtman DP (2006) Transporters expressed during grape berry (Vitis vinifera L.) development are associated with an increase in berry size and berry potassium accumulation. J Exp Bot 57:3209–3216. https://doi.org/10.1093/jxb/erl091
Desbrosses G, Kopka C, Ott T, Udvardi MK (2004) Lotus japonicus LjKUP is induced late during nodule development and encodes a potassium transporter of the plasma membrane. Mol Plant-Microbe Interact 17:789–797. https://doi.org/10.1094/MPMI.2004.17.7.789
Dreyer I, Uozumi N (2011) Potassium channels in plant cells. FEBS J 278:4293–4303. https://doi.org/10.1111/j.1742-4658.2011.08371.x
Garciadeblas B, Benito B, Rodríguez-Navarro A (2002) Molecular cloning and functional expression in bacteria of the potassium transporters CnHAK1 and CnHAK2 of the seagrass Cymodocea nodosa. Plant Mol Biol 50:623–633
Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, Michaux-Ferrière N, Thibaud JB, Sentenac H (1998) Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94:647–655
Gomez-Porras JL, Riaño-Pachón DM, Benito B, Haro R, Sklodowski K, Rodríguez-Navarro A, Dreyer I (2012) Phylogenetic analysis of K+ transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Front Plant Sci 3:167. https://doi.org/10.3389/fpls.2012.00167
Gupta AS, Berkowitz GA, Pier PA (1989) Maintenance of photosynthesis at low leaf water potential in wheat: role of potassium status and irrigation history. Plant Physiol 89:1358–1365
Hamamoto S, Marui J, Matsuoka K, Higashi K, Igarashi K, Nakagawa T, Kuroda T, Mori Y, Murata Y, Nakanishi Y, Maeshima M, Yabe I, Uozumi N (2008) Characterization of a tobacco TPK-type K+ channel as a novel tonoplast K+ channel using yeast tonoplasts. J Biol Chem 283:1911–1920. https://doi.org/10.1074/jbc.M708213200
Hamamoto S, Horie T, Hauser F, Deinlein U, Schroeder JI, Uozumi N (2015) HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr Opin Biotechnol 32:113–120. https://doi.org/10.1016/j.copbio.2014.11.025
Hedrich R (2012) Ion channels in plants. Physiol Rev 92:1777–1811. https://doi.org/10.1152/physrev.00038.2011
Hosy E, Vavasseur A, Mouline K, Dreyer I, Gaymard F, Poree F, Boucherez J, Lebaudy A, Bouchez D, Very AA, Simonneau T, Thibaud JB, Sentenac H (2003) The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc Natl Acad Sci U S A 100:5549–5554. https://doi.org/10.1073/pnas.0733970100
Hyun TK, Rim Y, Kim E, Kim J-S (2014) Genome-wide and molecular evolution analyses of the KT/HAK/KUP family in tomato (Solanum lycopersicum L.). Genes Genomics 36:365–374. https://doi.org/10.1007/s13258-014-0174-0
Isayenkov S, Isner J-C, Maathuis FJM (2011) Rice two-pore K+ channels are expressed in different types of vacuoles. Plant Cell 23:756–768. https://doi.org/10.1105/tpc.110.081463
Jain M, Misra G, Patel RK, Priya P, Jhanwar S, Khan AW, Shah N, Singh VK, Garg R, Jeena G, Yadav M, Kant C, Sharma P, Yadav G, Bhatia S, Tyagi AK, Chattopadhyay D (2013) A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J 74:715–729. https://doi.org/10.1111/tpj.12173
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282
Kato Y, Sakaguchi M, Mori Y, Saito K, Nakamura T, Bakker EP, Sato Y, Goshima S, Uozumi N (2001) Evidence in support of a four transmembrane-pore-transmembrane topology model for the Arabidopsis thaliana Na+/K+ translocating AtHKT1 protein, a member of the superfamily of K+ transporters. Proc Natl Acad Sci U S A 98:6488–6493. https://doi.org/10.1073/pnas.101556598
Koizumi N, Ujino T, Sano H, Chrispeels MJ (1999) Overexpression of a gene that encodes the first enzyme in the biosynthesis of asparagine-linked glycans makes plants resistant to tunicamycin and obviates the tunicamycin-induced unfolded protein response. Plant Physiol 121:353–361
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7 . 0 for bigger datasets brief communication. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lacombe B, Pilot G, Michard E, Gaymard F, Sentenac H, Thibaud JB (2000) A shaker-like K(+) channel with weak rectification is expressed in both source and sink phloem tissues of Arabidopsis. Plant Cell 12:837–851
Latz A, Becker D, Hekman M, Müller T, Beyhl D, Marten I, Eing C, Fischer A, Dunkel M, Bertl A, Rapp UR, Hedrich R (2007) TPK1, a Ca(2+)-regulated Arabidopsis vacuole two-pore K(+) channel is activated by 14-3-3 proteins. Plant J 52:449–459. https://doi.org/10.1111/j.1365-313X.2007.03255.x
Lebaudy A, Véry A-A, Sentenac H (2007) K+ channel activity in plants: genes, regulations and functions. FEBS Lett 581:2357–2366. https://doi.org/10.1016/j.febslet.2007.03.058
Lebaudy A, Vavasseur A, Hosy E, Dreyer I, Leonhardt N, Thibaud JB, Véry AA, Simonneau T, Sentenac H (2008) Plant adaptation to fluctuating environment and biomass production are strongly dependent on guard cell potassium channels. Proc Natl Acad Sci U S A 105:5271–5276. https://doi.org/10.1073/pnas.0709732105
Leigh RA (2001) Potassium homeostasis and membrane transport. J Plant Nutr Soil Sci 164:193–198. https://doi.org/10.1002/1522-2624(200104)164:2<193::AID-JPLN193>3.0.CO;2-7
Leonhardt N, Kwak JMJ, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16:596–615. https://doi.org/10.1105/tpc.019000.2
Lerouge P, Cabanes-Macheteau M, Rayon C, Fischette-Lainé AC, Gomord V, Faye L (1998) N-glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol Biol 38:31–48. https://doi.org/10.1023/A:1006012005654
Li W, Liu B, Yu L, Feng D, Wang H, Wang J (2009) Phylogenetic analysis, structural evolution and functional divergence of the 12-oxo-phytodienoate acid reductase gene family in plants. BMC Evol Biol 9:90. https://doi.org/10.1186/1471-2148-9-90
Li W-Y, Wang X, Li R, Li WQ, Chen KM (2014) Genome-wide analysis of the NADK gene family in plants. PLoS One 9:e101051. https://doi.org/10.1371/journal.pone.0101051
Liu K, Li L, Luan S (2006) Intracellular K+ sensing of SKOR, a Shaker-type K+ channel from Arabidopsis. Plant J 46:260–268. https://doi.org/10.1111/j.1365-313X.2006.02689.x
Maathuis FJM (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12:250–258. https://doi.org/10.1016/j.pbi.2009.04.003
Maathuis FJM, Filatov V, Herzyk P, C. Krijger G, B. Axelsen K, Chen S, Green BJ, Li Y, Madagan KL, Sánchez-Fernández R, Forde BG, Palmgren MG, Rea PA, Williams LE, Sanders D, Amtmann A (2003) Transcriptome analysis of root transporters reveals participation of multiple gene families in the response to cation stress. Plant J 35:675–692. https://doi.org/10.1046/j.1365-313X.2003.01839.x
Marcel D, Müller T, Hedrich R, Geiger D (2010) K+ transport characteristics of the plasma membrane tandem-pore channel TPK4 and pore chimeras with its vacuolar homologs. FEBS Lett 584:2433–2439. https://doi.org/10.1016/j.febslet.2010.04.038
Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJ, Sanders D, Harper JF, Tchieu J, Gribskov M, Persans MW, Salt DE, Kim SA, Guerinot ML (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126:1646–1667. https://doi.org/10.1104/pp.126.4.1646
Moshelion M, Becker D, Czempinski K, Mueller-Roeber B, Attali B, Hedrich R, Moran N (2002) Diurnal and circadian regulation of putative potassium channels in a leaf moving organ. Plant Physiol 128:634–642. https://doi.org/10.1104/pp.010549
Munro AW, Ritchie GY, Lamb AJ, Douglas RM, Booth IR (1991) The cloning and DNA sequence of the gene for the glutathione-regulated potassium-efflux system KefC of Escherichia coli. Mol Microbiol 5:607–616
Nieves-Cordones M, Martínez V, Benito B, Rubio F (2016) Comparison between Arabidopsis and rice for main pathways of K+ and Na+ uptake by roots. Front Plant Sci 7. https://doi.org/10.3389/fpls.2016.00992
Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran L-SP (2013) Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J Exp Bot 64:445–458. https://doi.org/10.1093/jxb/ers354
Pilot G, Gaymard F, Mouline K, Chérel I, Sentenac H (2003a) Regulated expression of Arabidopsis shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol Biol 51:773–787
Pilot G, Pratelli R, Gaymard F et al (2003b) Five-group distribution of the shaker-like K+ channel family in higher plants. J Mol Evol 56:418–434. https://doi.org/10.1007/s00239-002-2413-2
Platten JD, Cotsaftis O, Berthomieu P, Bohnert H, Davenport RJ, Fairbairn DJ, Horie T, Leigh RA, Lin HX, Luan S, Mäser P, Pantoja O, Rodríguez-Navarro A, Schachtman DP, Schroeder JI, Sentenac H, Uozumi N, Véry AA, Zhu JK, Dennis ES, Tester M (2006) Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 11:372–374. https://doi.org/10.1016/j.tplants.2006.06.001
Rasool S, Latef AAHA, Ahmad P (2015) Chickpea. In: Legumes under environmental stress. Wiley, Chichester, pp 67–79
Reintanz B, Szyroki A, Ivashikina N, Ache P, Godde M, Becker D, Palme K, Hedrich R (2002) AtKC1, a silent Arabidopsis potassium channel alpha-subunit modulates root hair K+ influx. Proc Natl Acad Sci U S A 99:4079–4084. https://doi.org/10.1073/pnas.052677799
Rubio F, Santa-Maria GE, Rodriguez-Navarro A (2000) Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol Plant 109:34–43. https://doi.org/10.1034/j.1399-3054.2000.100106.x
Rubio F, Alemán F, Nieves-Cordones M, Martínez V (2010) Studies on Arabidopsis athak5, atakt1 double mutants disclose the range of concentrations at which AtHAK5, AtAKT1 and unknown systems mediate K uptake. Physiol Plant 139:220–228. https://doi.org/10.1111/j.1399-3054.2010.01354.x
Schroeder JI, Ward JM, Gassmann W (1994) Perspectives on the physiology and structure of inward-rectifying K+ channels in higher plants: biophysical implications for K+ uptake. Annu Rev Biophys Biomol Struct 23:441–471. https://doi.org/10.1146/annurev.bb.23.060194.002301
Senn ME, Rubio F, Bañuelos MA, Rodríguez-Navarro A (2001) Comparative functional features of plant potassium HvHAK1 and HvHAK2 transporters. J Biol Chem 276:44563–44569. https://doi.org/10.1074/jbc.M108129200
Sharma T, Dreyer I, Riedelsberger J (2013) The role of K+ channels in uptake and redistribution of potassium in the model plant Arabidopsis thaliana. Front Plant Sci 4. https://doi.org/10.3389/fpls.2013.00224
Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci U S A 97:6896–6901. https://doi.org/10.1073/pnas.120170197
Singh KB, Ocampo B, Robertson LD (1998) Diversity for abiotic and biotic stress resistance in the wild annual Cicer species. Genet Resour Crop Evol 45:9–17. https://doi.org/10.1023/A:1008620002136
Strasser R (2016) Plant protein glycosylation. Glycobiology 26:926–939. https://doi.org/10.1093/glycob/cww023
Su H, Golldack D, Zhao C, Bohnert HJ (2002) The expression of HAK-type K(+) transporters is regulated in response to salinity stress in common ice plant. Plant Physiol 129:1482–1493. https://doi.org/10.1104/pp.001149
Tholema N, Bruggen MV d, Maser P et al (2005) All four putative selectivity filter glycine residues in KtrB are essential for high affinity and selective K+ uptake by the KtrAB system from Vibrio alginolyticus. J Biol Chem 280:41146–41154. https://doi.org/10.1074/jbc.M507647200
Vallejo AJ, ML P, GE S-M (2005) Expression of potassium-transporter coding genes, and kinetics of rubidium uptake, along a longitudinal root axis. Plant Cell Environ 28:850–862. https://doi.org/10.1111/j.1365-3040.2005.01334.x
van Geest M, Lolkema JS (2000) Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol Mol Biol Rev 64:13–33. https://doi.org/10.1128/MMBR.64.1.13-33.2000
Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, Cannon S, Baek J, Rosen BD, Tar'an B, Millan T, Zhang X, Ramsay LD, Iwata A, Wang Y, Nelson W, Farmer AD, Gaur PM, Soderlund C, Penmetsa RV, Xu C, Bharti AK, He W, Winter P, Zhao S, Hane JK, Carrasquilla-Garcia N, Condie JA, Upadhyaya HD, Luo MC, Thudi M, Gowda CLL, Singh NP, Lichtenzveig J, Gali KK, Rubio J, Nadarajan N, Dolezel J, Bansal KC, Xu X, Edwards D, Zhang G, Kahl G, Gil J, Singh KB, Datta SK, Jackson SA, Wang J, Cook DR (2013) Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 31:240–246. https://doi.org/10.1038/nbt.2491
Véry A-A, Sentenac H (2003) Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol 54:575–603. https://doi.org/10.1146/annurev.arplant.54.031902.134831
Véry A-A, Nieves-Cordones M, Daly M, Khan I, Fizames C, Sentenac H (2014) Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J Plant Physiol 171:748–769. https://doi.org/10.1016/j.jplph.2014.01.011
Voelker C, Gomez-Porras JL, Becker D, Hamamoto S, Uozumi N, Gambale F, Mueller-Roeber B, Czempinski K, Dreyer I (2010) Roles of tandem-pore K+ channels in plants—a puzzle still to be solved. Plant Biol 12:56–63. https://doi.org/10.1111/j.1438-8677.2010.00353.x
Walker DJ, R a L, Miller a J (1996) Potassium homeostasis in vacuolate plant cells. Proc Natl Acad Sci U S A 93:10510–10514
Wang Y-H, Garvin DF, Kochian LV (2002) Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol 130:1361–1370. https://doi.org/10.1104/pp.008854
Wang M, Zheng Q, Shen Q, Guo S (2013) The critical role of potassium in plant stress response. Int J Mol Sci 14:7370–7390. https://doi.org/10.3390/ijms14047370
Ward JM, Mäser P, Schroeder JI (2009) Plant ion channels: gene families, physiology, and functional genomics analyses. Annu Rev Physiol 71:59–82. https://doi.org/10.1146/annurev.physiol.010908.163204
Yang T, Zhang S, Hu Y, Wu F, Hu Q, Chen G, Cai J, Wu T, Moran N, Yu L, Xu G (2014) The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol 166:945–959. https://doi.org/10.1104/pp.114.246520
Funding
This study was funded by International Foundation for Science (IFS) (grant number no. C/5684-1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic Supplementary Material
Table S1
(DOCX 17 kb)
Table S2
(DOCX 21 kb)
Table S3
(DOCX 12 kb)
Figure S1
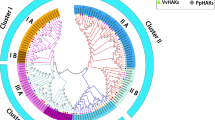
A) An unrooted phylogenetic tree of HKT family is generated via MEGA 7. The complete protein sequences of all members were aligned to generate the tree. Two subfamilies of HKTs (subfamily I and subfamily II) on the base of G or S residues are also shown in the tree. (B) Multiple sequence alignment of HKT family is shown. First P loop of various plant (Cicer arietinum L, Eucalyptus camaldulensis, Oryza sativa and Mesembryanthemum crystallinum) HKTs is compared with TrkH from Pseudomonas aeruginosa, P-loop of the Drosophila Shaker channel, Trk1from S. cerevisiae and to the KtrB from Vibrio alginolyticus, Alignment was done via ClustalX2. Conserved G residue is shown above the alignment. (JPG 202 kb)
Figure S2
(A) An unrooted phylogenetic tree of KUP/HAK/KT family is presented. The tree was generated using MEGA 7. The complete protein sequence of all members was aligned and TreeView program was used for visualization and analysis of graphical output. (B) Multiple sequence alignment of KUP/HAK/KT family is shown. Alignment was done via ClustalX2. Conserved motifs are shown above the alignment. Highly conserved residues are highlighted. (JPG 377 kb)
Figure S3
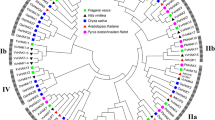
(A) An unrooted phylogenetic tree of KEA family is given. The tree was generated using Neighbor joining method via MEGA 7. TreeView program was used for visualization of graphical output. (B) Multiple sequence alignment of KEA family is shown. Alignment was done via ClustalX2. Conserved motifs are shown above the alignment. Highly conserved residues are highlighted. (JPG 348 kb)
Figure S4
(A) An unrooted phylogenetic tree of Shaker family is presented. The tree was generated using MEGA 7. The complete protein sequence of all members was aligned to generate the tree. Subfamilies of Shaker family are also shown in the tree. (B) Multiple sequence alignment of Shaker family is shown. Alignment was done via ClustalX2. TXXTXGYGD motif, a hallmark of K+ channels, is shown above the alignment . Highly conserved residues are highlighted in black. (JPG 447 kb)
Figure S5
(A) An unrooted phylogenetic tree of KCO family is given. The tree was generated using Neighbor joining method via MEGA 7. TreeView program was used for visualization and analysis of tree. (B)Multiple sequence alignment of KCO family is shown. Alignment was done via ClustalX2. Conserved motifs i.e. RSXpSX, necessary to interact with regulatory proteins, and TXXTXGYGD (a hallmark of K+ channels) are shown above the alignment. Highly conserved residues are highlighted in black. (JPG 220 kb)
Rights and permissions
About this article
Cite this article
Azeem, F., Ahmad, B., Atif, R.M. et al. Genome-Wide Analysis of Potassium Transport-Related Genes in Chickpea (Cicer arietinum L.) and Their Role in Abiotic Stress Responses. Plant Mol Biol Rep 36, 451–468 (2018). https://doi.org/10.1007/s11105-018-1090-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-018-1090-2