Abstract
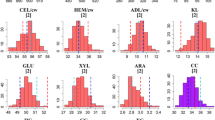
Genetic studies are ideal platforms for assessing the extent of genetic diversity, inferring the genetic architecture, and evaluating complex trait interrelations for cell wall compositional and bioconversion traits relevant to bioenergy applications. Through the characterization of a forage maize doubled haploid (DH) population, we indicate the substantial degree of highly heritable (h 2 > ~65 %) diversity in cell wall composition and bioconversion potential available within this important agronomic species. In addition to variation in lignin content, extensive genotypic diversity was found for the concentration and composition of hemicelluloses, the latter found to exert an influence on the recalcitrance of maize cell walls. Our results also demonstrate that forage maize harbors considerable variation for the release of cell wall glucose following pretreatment and enzymatic saccharification. In fact, the extent of variability observed for bioconversion efficiency (nearly 30 % between population extremes) greatly exceeded ranges reported in previous studies. In our population, a total of 52 quantitative trait loci (QTL) were detected for biomass compositional and bioconversion characters across 8 chromosomes. Noteworthy, from eight QTL related to bioconversion properties, five were previously unidentified and warrant further investigation. Ultimately, our results substantiate forage maize germplasm as a valid genetic resource for advancing cell wall degradability traits in bioenergy maize-breeding programs. However, since useful variation for cell wall traits is defined by QTL with “minor” effects (R 2 = ~10 %), cultivar development for bio-based applications will rely on advanced marker-assisted selection procedures centered on detecting and increasing the frequency of favorable QTL alleles in elite flint and dent germplasm.






Similar content being viewed by others
References
Bacovsky D (2010) How close are second-generation biofuels? Biofuels Bioprod Bioref 4(3):249–252
Saddler JN, Mabee WE, Simms R, Taylor M (2012) The biorefining story: progress in the commercialization of biomass-to-ethanol. In: Forests in development: a vital balance. Springer, pp 39-51
Wyman CE (2007) What is (and is not) vital to advancing cellulosic ethanol. Trends Biotechnol 25(4):153–157
Mosier N, Wyman C, Dale B, Elander R, Lee Y, Holtzapple M, Ladisch M (2005) Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol 96(6):673–686
Vermerris W, Saballos A, Ejeta G, Mosier NS, Ladisch MR, Carpita NC (2007) Molecular breeding to enhance ethanol production from corn and sorghum stover. Crop Sci 47(Supplement_3):S-142–S-153
Elander R, Dale B, Holtzapple M, Ladisch M, Lee YY, Mitchinson C, Saddler J, Wyman C (2009) Summary of findings from the Biomass Refining Consortium for Applied Fundamentals and Innovation (CAFI): corn stover pretreatment. Cellulose 16(4):649–659
Yang B, Wyman CE (2007) Pretreatment: the key to unlocking low–cost cellulosic ethanol. Biofuels Bioprod Bioref 1:26–40
Torres AF, van der Weijde T, Dolstra O, Visser RG, Trindade LM (2013) Effect of maize biomass composition on the optimization of dilute-acid pretreatments and enzymatic saccharification. BioEnergy Res 6(3):1038–1051
Carroll A, Somerville C (2009) Cellulosic biofuels. Annu Rev Plant Biol 60:165–182
Carpita N, Tierney M, Campbell M (2001) Molecular biology of the plant cell wall: searching for the genes that define structure, architecture and dynamics. Plant Mol Biol 47:1–5
Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6(11):850–861
Carpita NC, McCann MC (2008) Maize and sorghum: genetic resources for bioenergy grasses. Trends Plant Sci 13(8):415–420
Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25(7):759–761
Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M, Chen F, Foston M, Ragauskas A, Bouton J, Dixon RA, Wang Z-Y (2011) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci U S A 108(9):3803–3808
Jung JH, Fouad WM, Vermerris W, Gallo M, Altpeter F (2012) RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol J 10(9):1067–1076
Dien B, Sarath G, Pedersen J, Sattler S, Chen H, Funnell-Harris D, Nichols N, Cotta M (2009) Improved sugar conversion and ethanol yield for forage sorghum (Sorghum bicolor L. Moench) lines with reduced lignin contents. BioEnergy Res 2(3):153–164
Lorenzana RE, Lewis MF, Jung HJG, Bernardo R (2010) Quantitative trait loci and trait correlations for maize stover cell wall composition and glucose release for cellulosic ethanol. Crop Sci 50(2):541–555
Saballos A, Vermerris W, Rivera L, Ejeta G (2008) Allelic association, chemical characterization and saccharification properties of brown midrib mutants of sorghum (Sorghum bicolor (L.) Moench). BioEnergy Res 1(3–4):193–204
Sarath G, Dien B, Saathoff AJ, Vogel KP, Mitchell RB, Chen H (2011) Ethanol yields and cell wall properties in divergently bred switchgrass genotypes. Bioresour Technol 102(20):9579–9585
Torres AF, Visser RG, Trindade LM (2013) Bioethanol from maize cell walls: genes, molecular tools and breeding prospects. GCB Bioenergy. doi:10.1111/gcbb.12164
Weijde T, Alvim Kamei CL, Torres AF, Vermerris W, Dolstra O, Visser RGF, Trindade LM (2013) The potential of C4 grasses for cellulosic biofuel production. Front Plant Sci 4:107. doi:10.3389/fpls.2013.00107
Lorenz A, Coors J, Hansey C, Kaeppler S, De Leon N (2010) Genetic analysis of cell wall traits relevant to cellulosic ethanol production in maize (L.). Crop Sci 50(3):842–852
Lorenz AJ, Coors JG, de Leon N, Wolfrum EJ, Hames BR, Sluiter AD, Weimer PJ (2009) Characterization, genetic variation, and combining ability of maize traits relevant to the production of cellulosic ethanol. Crop Sci 49(1):85–98
Lauer J, Coors J, Flannery P (2001) Forage yield and quality of corn cultivars developed in different eras. Crop Sci 41(5):1449–1455
Argillier O, Barrière Y, Lila M, Jeanneteau F, Gélinet K, Ménanteau V (1996) Genotypic variation in phenolic components of cell-walls in relation to the digestibility of maize stalks. Agronomie 16(2):123–130
Barrière Y, Thomas J, Denoue D (2008) QTL mapping for lignin content, lignin monomeric composition, p-hydroxycinnamate content, and cell wall digestibility in the maize recombinant inbred line progeny F838× F286. Plant Sci 175(4):585–595
Barrière Y, Méchin V, Lafarguette F, Manicacci D, Guillon F, Wang H, Lauressergues D, Pichon M, Bosio M, Tatout C (2009) Toward the discovery of maize cell wall genes involved in silage quality and capacity to biofuel production. Maydica 54(2):161–198
Barrière Y, Charcosset A, Denoue D, Madur D, Bauland C, Laborde J (2010) Genetic variation for lignin content and cell wall digestibility in early maize lines derived from ancient landraces. Maydica 55(1):65–74
Fontaine A-S, Bout S, Barrière Y, Vermerris W (2003) Variation in cell wall composition among forage maize (Zea mays L.) inbred lines and its impact on digestibility: analysis of neutral detergent fiber composition by pyrolysis-gas chromatography-mass spectrometry. J Agric Food Chem 51(27):8080–8087
Marita JM, Vermerris W, Ralph J, Hatfield RD (2003) Variations in the cell wall composition of maize brown midrib mutants. J Agric Food Chem 51(5):1313–1321
Marvin HJP, Krechting CF, Van Loo EN, Snijders CHA, Dolstra O (1995) Relationship between stalk cell wall digestibility and fibre composition in maize. J Sci Food Agric 69(2):215–221
Méchin V, Argillier O, Hébert Y, Guingo E, Moreau L, Charcosset A, Barrière Y (2001) Genetic analysis and QTL mapping of cell wall digestibility and lignification in silage maize. Crop Sci 41(3):690–697
Dolstra O, Medema J (1990) An effective screening method for genetic improvement of cell-wall digestibility in forage maize. In: Proceedings 15th congress maize and sorghum section of Eucarpia. pp 4-8
Dolstra O, Medema J, De Jong A (1992) Genetic improvement of cell-wall digestibility in forage maize (Zea mays L.). I. Performance of inbred lines and related hybrids. Euphytica 65(3):187–194
Goering H, Van Soest PJ (1970) Forage fiber analyses (apparatus, reagents, procedures, and some applications), vol 379. US Agricultural Research Service, Washington
Englyst HN, Cummings JH (1984) Simplified method for the measurement of total non-starch polysaccharides by gas-liquid chromatography of constituent sugars as alditol acetates. Analyst 109(7):937–942
Selig M, Weiss N, Ji Y (1996) Enzymatic saccharification of lignocellulosic biomass; LAP-009 NREL analytical procedure. National Renewable Energy Laboratory, Golden
Dewey DR, Lu K (1959) A correlation and path-coefficient analysis of components of crested wheatgrass seed production. Agron J 51(9):515–518
Jung H-JG, Bernardo R (2012) Comparison of cell wall polysaccharide hydrolysis by a dilute acid/enzymatic saccharification process and rumen microorganisms. BioEnergy Res 5(2):319–329
Méchin V, Argillier O, Menanteau V, Barrière Y, Mila I, Pollet B, Lapierre C (2000) Relationship of cell wall composition to in vitro cell wall digestibility of maize inbred line stems. J Sci Food Agric 80(5):574–580
Courtial A, Méchin V, Reymond M, Grima-Pettenati J, Barrière Y (2014) Colocalizations between several QTLs for cell wall degradability and composition in the F288× F271 early maize RIL progeny raise the question of the nature of the possible underlying determinants and breeding targets for biofuel capacity. BioEnergy Res 7:142–156
Riboulet C, Lefevre B, Denoue D, Barrière Y (2008) Genetic variation in maize cell wall for lignin content, lignin structure, p-hydroxycinnamic acid content, and digestibility in set of 19 lines at silage harvest maturity. Maydica 53:11–19
Vermerris W, Sherman DM, McIntyre LM (2010) Phenotypic plasticity in cell walls of maize brown midrib mutants is limited by lignin composition. J Exp Bot 61(9):2479–2490
Grabber J, Hatfield R, Ralph J, Zon J, Amrhein N (1995) Ferulate cross-linking in cell walls isolated from maize cell suspensions. Phytochemistry 40:1077–1082
Grabber J, Ralph J, Hatfield R (2000) Cross-linking of maize walls by ferulate dimerization and incorporation into lignin. J Agric Food Chem 48:6106–6113
Grabber JH, Ralph J, Lapierre C, Barrière Y (2004) Genetic and molecular basis of grass cell-wall degradability. I. Lignin–cell wall matrix interactions. C R Biol 327(5):455–465
Mortimer JC, Miles GP, Brown DM, Zhang Z, Segura MP, Weimar T, Yu X, Seffen KA, Stephens E, Turner SR, Dupree P (2010) Absence of branches from xylan in Arabidopsis gux mutants reveals potential for simplification of lignocellulosic biomass. Proc Natl Acad Sci U S A 107(40):17409–17414
Bromley JR, Busse-Wicher M, Tryfona T, Mortimer JC, Zhang Z, Brown DM, Dupree P (2013) GUX1 and GUX2 glucuronyltransferases decorate distinct domains of glucuronoxylan with different substitution patterns. Plant J 74(3):423–434
Kabel MA, van den Borne H, Vincken JP, Voragen AGJ, Schols HA (2007) Structural differences of xylans affect their interaction with cellulose. Carbohydr Polym 69(1):94–105
Grabber JH, Hatfield RD, Ralph J (1998) Diferulate cross-links impede the enzymatic degradation of non-lignified maize walls. J Sci Food Agric 77(2):193–200
Grabber JH, Mertens DR, Kim H, Funk C, Lu F, Ralph J (2009) Cell wall fermentation kinetics are impacted more by lignin content and ferulate cross-linking than by lignin composition. J Sci Food Agric 89(1):122–129
Grabber JH, Ralph J, Hatfield RD (1998) Ferulate cross-links limit the enzymatic degradation of synthetically lignified primary walls of maize. J Agric Food Chem 46(7):2609–2614
Penning BW, Hunter CT, Tayengwa R, Eveland AL, Dugard CK, Olek AT, Vermerris W, Koch KE, McCarty DR, Davis MF (2009) Genetic resources for maize cell wall biology. Plant Physiol 151(4):1703–1728
Wallace J, Larsson S, Buckler E (2013) Entering the second century of maize quantitative genetics. Heredity 112:30–38
Yin X, Struik PC, Kropff MJ (2004) Role of crop physiology in predicting gene-to-phenotype relationships. Trends Plant Sci 9(9):426–432
Melchinger AE, Utz HF, Schön CC (1998) Quantitative trait locus (QTL) mapping using different testers and independent population samples in maize reveals low power of QTL detection and large bias in estimates of QTL effects. Genetics 149(1):383–403
Peiffer JA, Flint-Garcia SA, De Leon N, McMullen MD, Kaeppler SM, Buckler ES (2013) The genetic architecture of maize stalk strength. PLoS ONE 8(6):e67066
Courtial A, Thomas J, Reymond M, Méchin V, Grima-Pettenati J, Barrière Y (2013) Targeted linkage map densification to improve cell wall related QTL detection and interpretation in maize. Theor Appl Genet 126(5):1151–1165
Riedelsheimer C, Czedik-Eysenberg A, Grieder C, Lisec J, Technow F, Sulpice R, Altmann T, Stitt M, Willmitzer L, Melchinger AE (2012) Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat Genet 44(2):217–220
Moose SP, Mumm RH (2008) Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol 147(3):969–977
Bennetzen J, Hake S, Vermerris W (2009) Cell wall biosynthetic genes of maize and their potential for bioenergy production. In: Bennetzen J, Hake S (eds) Handbook of maize. Springer, New York, pp 741–767
Carrier M, Joubert J-E, Danje S, Hugo T, Görgens J, Knoetze JH (2013) Impact of the lignocellulosic material on fast pyrolysis yields and product quality. Bioresour Technol 150:129–138
Burhenne L, Messmer J, Aicher T, Laborie M-P (2013) The effect of the biomass components lignin, cellulose and hemicellulose on TGA and fixed bed pyrolysis. J Anal Appl Pyrol 101:177–184
Vandenbrink JP, Hilten RN, Das K, Paterson AH, Feltus FA (2012) Analysis of crystallinity index and hydrolysis rates in the bioenergy crop Sorghum bicolor. BioEnergy Res 5(2):387–397
Zhang W, Yi Z, Huang J, Li F, Hao B, Li M, Hong S, Lv Y, Sun W, Ragauskas A (2013) Three lignocellulose features that distinctively affect biomass enzymatic digestibility under NaOH and H2S04 pretreatments in Miscanthus. Bioresour Technol 130:30–37
Harris D, Stork J, Debolt S (2009) Genetic modification in cellulose–synthase reduces crystallinity and improves biochemical conversion to fermentable sugar. GCB Bioenergy 1(1):51–61
Fornalé S, Capellades M, Encina A, Wang K, Irar S, Lapierre C, Ruel K, Joseleau J-P, Berenguer J, Puigdomènech P (2012) Altered lignin biosynthesis improves cellulosic bioethanol production in transgenic maize plants down-regulated for cinnamyl alcohol dehydrogenase. Mol Plant 5(4):817–830
Zeng M, Ximenes E, Ladisch MR, Mosier NS, Vermerris W, Huang CP, Sherman DM (2012) Tissue–specific biomass recalcitrance in corn stover pretreated with liquid hot–water: enzymatic hydrolysis (part 1). Biotechnol Bioeng 109(2):390–397
Lam M, Martinez Y, Barbier O, Jauneau A, Pichon M, Barrière Y, Motto M (2013) Maize cell wall degradability, from whole plant to tissue level: different scales of complexity. Maydica 58(2013):103–110
Lam MSWQ, Martinez Y, Barbier O, Jauneau A, Balzergue S, Huguet S, Pollet B, Méchin V, Guillon F, Robert P (2013) Combined approaches provide an anatomical and transcriptomic fingerprint of maize cell wall digestibility. Maydica 58(2013):274–287
Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM (2011) Genome–wide atlas of transcription during maize development. Plant J 66(4):553–563
Knox JP (2008) Revealing the structural and functional diversity of plant cell walls. Curr Opin Plant Biol 11(3):308–313
Roussel V, Gibelin C, Fontaine A, Barriere Y (2002) Genetic analysis in recombinant inbred lines of early dent forage maize. II. QTL mapping for cell wall constituents and cell wall digestibility from per se value and top cross experiments. Maydica 47(1):9–20
Barrière Y, Méchin V, Lefevre B, Maltese S (2012) QTLs for agronomic and cell wall traits in a maize RIL progeny derived from a cross between an old Minnesota13 line and a modern Iodent line. Theor Appl Genet 125(3):531–549
Truntzler M, Barrière Y, Sawkins M, Lespinasse D, Betran J, Charcosset A, Moreau L (2010) Meta-analysis of QTL involved in silage quality of maize and comparison with the position of candidate genes. Theor Appl Genet 121(8):1465–1482
Acknowledgments
We gratefully acknowledge Genencor International B.V. for kindly supplying us with their cellulolytic enzyme cocktails used in this study. Within the framework of the Carbohydrate Competence Centre, this research has been financially supported by the European Union, the European Regional Development Fund, and the Northern Netherlands Provinces (Samenwerkingsverband Noord-Nederland), KOERS NOORD.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
(PDF 35 kb)
Fig. S1
Survey of QTLs identified for biomass compositional and bioconversion characters in a forage maize DH population. Colored bars indicate total number of QTLs identified per trait. Circular points refer to the cumulative explained variance of all identified QTLs for a given trait, as a proportion of observed heritable variation (h 2) (PPTX 54 kb)
Rights and permissions
About this article
Cite this article
Torres, A.F., Noordam-Boot, C.M.M., Dolstra, O. et al. Cell Wall Diversity in Forage Maize: Genetic Complexity and Bioenergy Potential. Bioenerg. Res. 8, 187–202 (2015). https://doi.org/10.1007/s12155-014-9507-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-014-9507-8