Abstract
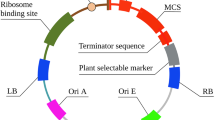
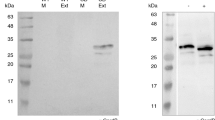
The natural source of chymosin, a key enzyme in the dairy industry, is insufficient for rapidly growing cheese industries. Large-scale production of recombinant proteins in heterologous hosts provides an efficient alternative solution. Here, the codon-optimized synthetic prochymosin gene, which has a CAI index of 0.926, was subcloned from a cloning vector (pUC57-bCYM) into the pBI121 vector, resulting in the construct named pBI121-bCYM. CAI ranges from 0 to 1 and higher CAI improves gene expression in heterologous hosts. The overexpression of the prochymosin gene was under the control of constitutive CaMV 35S promoter and NOS terminator and was transferred into the tobacco via A. tumefaciens strain LBA4404. Explant type, regeneration method, inoculation temperature, cell density (OD600) of Agrobacterium for inoculation, and acetosyringone concentration were leaf explants, direct somatic embryogenesis, 19 °C, 0.1, and 100 µM, respectively. The successful integration and expression of the prochymosin gene, along with the bioactivity of recombinant chymosin, were confirmed by PCR, RT-PCR, and milk coagulation assay, respectively. Overall, this study reports the first successful overexpression of the codon-optimized prochymosin form of the bovine chymosin enzyme in the tobacco via indirect transformation. Production of recombinant bovine chymosin in plants can be an easy-to-scale-up, safe, and inexpensive platform.




Similar content being viewed by others
Data Availability
Not available.
References
Alavi, F., & Momen, S. (2020). Aspartic proteases from thistle flowers: Traditional coagulants used in the modern cheese industry. International Dairy Journal, 107, 104709.
Kumar, A., Grover, S., Sharma, J., & Batish, V. (2010). Chymosin and other milk coagulants: Sources and biotechnological interventions. Critical Reviews in Biotechnology, 30, 243–258.
Shah, M. A., Mir, S. A., & Paray, M. A. (2014). Plant proteases as milk-clotting enzymes in cheesemaking: A review. Dairy Science & Technology, 94, 5–16.
Fernández-Salguero, J., Prados, F., Calixto, F., Vioque, M., Sampaio, P., & Tejada, L. (2003). Use of recombinant cyprosin in the manufacture of ewe’s milk cheese. Journal of Agricultural and Food Chemistry, 51, 7426–7430.
Menacho-Melgar, R., Ye, Z., Moreb, E. A., Yang, T., Efromson, J. P., Decker, J. S., Wang, R., & Lynch, M. D. (2020). Scalable, two-stage, autoinduction of recombinant protein expression in E. coli utilizing phosphate depletion. Biotechnology and Bioengineering, 117, 2715–2727.
Packiam, K. A. R., Ramanan, R. N., Ooi, C. W., Krishnaswamy, L., & Tey, B. T. (2020). Stepwise optimization of recombinant protein production in Escherichia coli utilizing computational and experimental approaches. Applied Microbiology and Biotechnology, 104, 3253–3266.
Pijlman, G. P., Grose, C., Hick, T. A., Breukink, H. E., van den Braak, R., Abbo, S. R., Geertsema, C., van Oers, M. M., Martens, D. E., & Esposito, D. (2020). Relocation of the attTn7 transgene insertion site in bacmid DNA enhances baculovirus genome stability and recombinant protein expression in insect cells. Viruses, 12, 1448.
Grose, C., Putman, Z., & Esposito, D. (2021). A review of alternative promoters for optimal recombinant protein expression in Baculovirus-infected insect cells. Protein Expression and Purification, 186, 105924.
Pourcel, L., Buron, F., Arib, G., Le Fourn, V., Regamey, A., Bodenmann, I., Girod, P. A., & Mermod, N. (2020). Influence of cytoskeleton organization on recombinant protein expression by CHO cells. Biotechnology and Bioengineering, 117, 1117–1126.
Frei, T., Cella, F., Tedeschi, F., Gutiérrez, J., Stan, G.-B., Khammash, M., & Siciliano, V. (2020). Characterization and mitigation of gene expression burden in mammalian cells. Nature Communications, 11, 1–14.
Ntana, F., Mortensen, U. H., Sarazin, C., & Figge, R. (2020). Aspergillus: A powerful protein production platform. Catalysts, 10, 1064.
Liu, Y., Li, Y., Tong, S., Yuan, M., Wang, X., Wang, J., & Fan, Y. (2020). Expression of a Beauveria bassiana chitosanase (BbCSN-1) in Pichia pastoris and enzymatic analysis of the recombinant protein. Protein Expression and Purification, 166, 105519.
Burnett, M. J., & Burnett, A. C. (2020). Therapeutic recombinant protein production in plants: Challenges and opportunities. Plants, People, Planet, 2, 121–132.
Shanmugaraj, B., I Bulaon, C. J., & Phoolcharoen, W. (2020). Plant molecular farming: a viable platform for recombinant biopharmaceutical production. Plants, 9, 842.
Rebelo, B. A., Santos, R. B., Ascenso, O. S., Nogueira, A. C., Lousa, D., Abranches, R., & Ventura, M. R. (2020). Synthesis and biological effects of small molecule enhancers for improved recombinant protein production in plant cell cultures. Bioorganic Chemistry, 94, 103452.
Fajardo, C., De Donato, M., Carrasco, R., Martínez-Rodríguez, G., Mancera, J. M., & Fernández-Acero, F. J. (2020). Advances and challenges in genetic engineering of microalgae. Reviews in Aquaculture, 12, 365–381.
Tran, N. T., & Kaldenhoff, R. (2020). Achievements and challenges of genetic engineering of the model green alga Chlamydomonas reinhardtii. Algal Research, 50, 101986.
Mohanty, A. K., Mukhopadhyay, U. K., Grover, S., & Batish, V. K. (1999). Bovine chymosin: Production by rDNA technology and application in cheese manufacture. Biotechnology Advances, 17, 205–217.
Gilliland, G. L., Winborne, E. L., Nachman, J., & Wlodawer, A. (1990). The three-dimensional structure of recombinant bovine chymosin at 2.3 Å resolution. Proteins: Structure. Function, and Bioinformatics, 8, 82–101.
Noseda, D. G., Recúpero, M. N., Blasco, M., Ortiz, G. E., & Galvagno, M. A. (2013). Cloning, expression and optimized production in a bioreactor of bovine chymosin B in Pichia (Komagataella) pastoris under AOX1 promoter. Protein Expression and Purification, 92, 235–244.
Wei, Z.-Y., Zhang, Y.-Y., Wang, Y.-P., Fan, M.-X., Zhong, X.-F., Xu, N., Lin, F., & Xing, S.-C. (2016). Production of bioactive recombinant bovine chymosin in tobacco plants. International Journal of Molecular Sciences, 17, 624.
Fischer, R., Stoger, E., Schillberg, S., Christou, P., & Twyman, R. M. (2004). Plant-based production of biopharmaceuticals. Current Opinion in Plant Biology, 7, 152–158.
Sindarovska, Y. R., Gerasymenko, I. M., Sheludko, Y. V., Olevinskaya, Z. M., Spivak, N. Y., & Kuchuk, N. V. (2010). Production of human interferon alfa 2b in plants of Nicotiana excelsior by Agrobacterium-mediated transient expression. Tsitologiia i Genetika, 44, 60–64.
Larrick, J. W., & Thomas, D. W. (2001). Producing proteins in transgenic plants and animals. Current Opinion in Biotechnology, 12, 411–418.
Azizi-Dargahlou, S., & Ahmadabadi, M. (2022). Antimicrobial peptides and their heterologous production in plant systems. New Cellular and Molecular Biotechnology Journal, 12, 9–22.
Norkiene, M., & Gedvilaite, A. (2012). Influence of codon bias on heterologous production of human papillomavirus type 16 major structural protein L1 in yeast. The Scientific World Journal. https://doi.org/10.1100/2012/979218
Elena, C., Ravasi, P., Castelli, M. E., Peirú, S., & Menzella, H. G. (2014). Expression of codon optimized genes in microbial systems: Current industrial applications and perspectives. Frontiers in Microbiology, 5, 21.
Sharp, P. M., & Li, W.-H. (1987). The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Research, 15, 1281–1295.
Puigbò, P., Bravo, I. G., & Garcia-Vallve, S. (2008). CAIcal: A combined set of tools to assess codon usage adaptation. Biology Direct, 3, 1–8.
Azizi Dargahlou, S., Ahmadabadi, M., & Valizadeh Kamran, R. (2022). Codon optimization and cloning of bovine prochymosin gene for proper expression in tobacco plant. Genetic Engineering and Biosafety Journal, 10, 225–236.
Azizi-Dargahlou, S., & Pouresmaeil, M. (2023). Agrobacterium tumefaciens-Mediated Plant Transformation: A Review. Molecular Biotechnology. https://doi.org/10.1007/s12033-023-00788-x
Li, S., Cong, Y., Liu, Y., Wang, T., Shuai, Q., Chen, N., Gai, J., & Li, Y. (2017). Optimization of Agrobacterium-mediated transformation in soybean. Frontiers in Plant Science, 8, 246.
Tiwari, M., Mishra, A. K., & Chakrabarty, D. (2022). Agrobacterium-mediated gene transfer: Recent advancements and layered immunity in plants. Planta, 256, 37.
Shreni Agrawal, E. R. (2022). A Review: Agrobacterium-mediated gene transformation to increase plant productivity. Journal of Phytopharmatics, 11, 111–117.
Che, P., Anand, A., Wu, E., Sander, J. D., Simon, M. K., Zhu, W., Sigmund, A. L., Zastrow-Hayes, G., Miller, M., & Liu, D. (2018). Developing a flexible, high-efficiency Agrobacterium-mediated sorghum transformation system with broad application. Plant Biotechnology Journal, 16, 1388–1395.
Mohammed, S., Abd Samad, A., & Rahmat, Z. (2019). Agrobacterium-mediated transformation of rice: Constraints and possible solutions. Rice Science, 26, 133–146.
Anand, A., Bass, S. H., Wu, E., Wang, N., McBride, K. E., Annaluru, N., Miller, M., Hua, M., & Jones, T. J. (2018). An improved ternary vector system for Agrobacterium-mediated rapid maize transformation. Plant Molecular Biology, 97, 187–200.
Song, G.-Q., Prieto, H., & Orbovic, V. (2019). Agrobacterium-mediated transformation of tree fruit crops: Methods, progress, and challenges. Frontiers in Plant Science, 10, 226.
Veillet, F., Perrot, L., Chauvin, L., Kermarrec, M.-P., Guyon-Debast, A., Chauvin, J.-E., Nogué, F., & Mazier, M. (2019). Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. International Journal of Molecular Sciences, 20, 402.
Javied, M., Ashfaq, N., Haider, M., Fatima, F., Ali, Q., Ali, A., & Malik, A. (2021). Agrobacterium-mediated transformation of cotton (Gossypium hirsutum L.) using dmo gene for enhanced tolerance against dicamba pesticide. Biological and Clinical Sciences Research Journal, 2021, e009–e009.
Jiang, H., Meng, F., Lu, D., Chen, Y., Luo, G., Chen, Y., Chen, J., Chen, C., Zhang, X., & Su, D. (2022). High-throughput fast cloning technology: A low-cost method for parallel cloning. PLoS ONE, 17, e0273873.
Engebrecht, J., Brent, R., & Kaderbhai, M. A. (1991). Minipreps of plasmid DNA. Current Protocols in Molecular Biology, 15, 1.6.1-1.6.10.
Murashige, T., & Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum, 15, 473–497.
Fu, Q., Li, C., Tang, M., Tao, Y.-B., Pan, B.-Z., Zhang, L., Niu, L., He, H., Wang, X., & Xu, Z.-F. (2015). An efficient protocol for Agrobacterium-mediated transformation of the biofuel plant Jatropha curcas by optimizing kanamycin concentration and duration of delayed selection. Plant Biotechnology Reports, 9, 405–416.
Yadav, M., Chaudhary, D., Sainger, M., & Jaiwal, P. K. (2010). Agrobacterium tumefaciens-mediated genetic transformation of sesame (Sesamum indicum L.). Plant Cell. Tissue and Organ Culture (PCTOC), 103, 377–386.
Salas, M., Park, S., Srivatanakul, M., & Smith, R. (2001). Temperature influence on stable T-DNA integration in plant cells. Plant Cell Reports, 20, 701–705.
Mortensen, S., Cole, L. F., Bernal-Franco, D., Sathitloetsakun, S., Cram, E. J., & Lee-Parsons, C. W. (2022). EASI transformation protocol: An agrobacterium-mediated transient transformation protocol for Catharanthus roseus seedlings. Catharanthus roseus: Methods and protocols (pp. 249–262). Springer.
Kauser, N., Khan, S., Mohammadi, A., Ghareyazil, B., Uliaie, E. D., & Darvishrohani, B. (2016). Agrobactrium mediated transformation and direct shoot regeneration in Iranian Tomato (Solanum lycopersicum L.) cultivar Falat-CH. Pakistan Journal of Botany, 48, 2489–2498.
Muppala, S., Gudlavalleti, P. K., Dasari, P., Pagidoju, S., Malireddy, K. R., & Puligundla, S. K. (2022). Agrobacterium mediated transformation of ABA biosynthetic pathway coding genes for enhanced drought tolerance in Nicotiana tabacum. Journal of Pharmacognosy and Phytochemistry, 11, 244–249.
Solís-Ramos, L. Y., Ortiz-Pavón, J. C., Andrade-Torres, A., Porras-Murillo, R., Angulo, A. B., & Serna, ECdl. (2019). Agrobacterium tumefaciens-mediated transformation of common bean (Phaseolus vulgaris) var Brunca. Revista de Biología Tropical, 67, 83–94.
Abdallah, N. A., Shah, D., Abbas, D., & Madkour, M. (2010). Stable integration and expression of a plant defensin in tomato confers resistance to fusarium wilt. GM Crops, 1, 344–350.
Li, J., Wang, S., Yu, J., Wang, L., & Zhou, S. (2013). A modified CTAB protocol for plant DNA extraction. Chinese Bulletin of Botany, 48, 72.
Azizi-Dargahlou, S., Ahmadabadi, M., & Valizadeh Kamran, R. (2023). Biolistic transformation and expression of functional chymosin from a codon-optimized synthetic bovine gene in tobacco Plants. Journal of Medicinal plants and By-product, 12, 209–215.
Liu, W.-G., Wang, Y.-P., Zhang, Z.-J., Wang, M., Lv, Q.-X., Liu, H.-W., Meng, L.-C., & Lu, M. (2017). Generation and characterization of caprine chymosin in corn seed. Protein Expression and Purification, 135, 78–82.
Noor, F., Ashfaq, U. A., Bakar, A., Qasim, M., Masoud, M. S., Alshammari, A., Alharbi, M., & Riaz, M. S. (2023). Identification and characterization of codon usage pattern and influencing factors in HFRS-causing hantaviruses. Frontiers in Immunology, 14, 1131647.
Fu, H., Liang, Y., Zhong, X., Pan, Z., Huang, L., Zhang, H., Xu, Y., Zhou, W., & Liu, Z. (2020). Codon optimization with deep learning to enhance protein expression. Scientific Reports, 10, 17617.
Vasbinder, A., Rollema, H., Bot, A., & De Kruif, C. (2003). Gelation mechanism of milk as influenced by temperature and pH; studied by the use of transglutaminase cross-linked casein micelles. Journal of Dairy Science, 86, 1556–1563.
Vallejo, J. A., Ageitos, J. M., Poza, M., & Villa, T. G. (2008). Cloning and expression of buffalo active chymosin in Pichia pastoris. Journal of Agricultural and Food Chemistry, 56, 10606–10610.
Al-Zoreky, N. S., & Almathen, F. S. (2021). Using recombinant camel chymosin to make white soft cheese from camel milk. Food Chemistry, 337, 127994.
Ersöz, F., & İnan, M. (2019). Large-scale production of yak (Bos grunniens) chymosin A in Pichia pastoris. Protein Expression and Purification, 154, 126–133.
Aboulnaga, E. (2019). Cloning and expression of camel pro-chymosin encoding gene in E. coli and characterization of the obtained active enzyme. Journal of Food and Dairy Sciences, 10, 71–78.
Tian, L., & Sun, S. S. (2011). A cost-effective ELP-intein coupling system for recombinant protein purification from plant production platform. PLoS ONE, 6, e24183.
Park, K. Y., & Wi, S. J. (2016). Potential of plants to produce recombinant protein products. Journal of Plant Biology, 59, 559–568.
Hood, E. E., & Howard, J. A. (2014). Commercial plant-produced recombinant avidin. Commercial plant-produced recombinant protein products: Case studies (pp. 15–25). Springer.
Bailey, M., Woodard, S., Callaway, E., Beifuss, K., Magallanes-Lundback, M., Lane, J., Horn, M., Mallubhotla, H., Delaney, D., & Ward, M. (2004). Improved recovery of active recombinant laccase from maize seed. Applied Microbiology and Biotechnology, 63, 390–397.
Gustafsson, C., Govindarajan, S., & Minshull, J. (2004). Codon bias and heterologous protein expression. Trends in Biotechnology, 22, 346–353.
Hershberg, R., & Petrov, D. A. (2008). Selection on codon bias. Annual Review of Genetics, 42, 287–299.
Espinoza-Molina, J. A., Acosta-Muniz, C. H., Sepulveda, D. R., Zamudio-Flores, P. B., & Rios-Velasco, C. (2016). Codon optimization of the “Bos taurus chymosin" gene for the production of recombinant chymosin in Pichia pastoris. Molecular Biotechnology, 58, 657–664.
Gouy, M., & Gautier, C. (1982). Codon usage in bacteria: Correlation with gene expressivity. Nucleic Acids Research, 10, 7055–7074.
Duret, L., & Mouchiroud, D. (1999). Expression pattern and surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proceedings of the National Academy of Sciences USA, 96, 4482–4487.
Hu, H., Dong, B., Fan, X., Wang, M., Wang, T., & Liu, Q. (2023). Mutational bias and natural selection driving the synonymous codon usage of single-exon genes in rice (Oryza sativa L.). Rice, 16, 1–13.
Feng, Z., Zhang, L., Han, X., & Zhang, Y. (2010). Codon optimization of the calf prochymosin gene and its expression in Kluyveromyces lactis. World Journal of Microbiology and Biotechnology, 26, 895–901.
Luo, F., Jiang, W. H., Yang, Y. X., Li, J., & Jiang, M. F. (2016). Cloning and expression of yak active chymosin in Pichia pastoris. Asian-Australasian Journal of Animal Sciences, 29, 1363.
Cardoza, R., Gutiérrez, S., Ortega, N., Colina, A., Casqueiro, J., & Martín, J. (2003). Expression of a synthetic copy of the bovine chymosin gene in Aspergillus awamori from constitutive and pH-regulated promoters and secretion using two different pre-pro sequences. Biotechnology and Bioengineering, 83, 249–259.
Khelifa, M., Massé, D., Blanc, S., & Drucker, M. (2010). Evaluation of the minimal replication time of Cauliflower mosaic virus in different hosts. Virology, 396, 238–245.
Krasnyanski, S. F., Sandhu, J., Domier, L. L., Buetow, D. E., & Korban, S. S. (2001). Effect of an enhanced CaMV 35S promoter and a fruit-specific promoter on uida gene expression in transgenic tomato plants. In Vitro Cellular & Developmental Biology-Plant, 37, 427–433.
Tepfer, M., Gaubert, S., Leroux-Coyau, M., Prince, S., & Houdebine, L.-M. (2004). Transient expression in mammalian cells of transgenes transcribed from the Cauliflower mosaic virus 35S promoter. Environmental Biosafety Research, 3, 91–97.
Sun, L., Cai, H., Xu, W., Hu, Y., & Lin, Z. (2002). CaMV 35S promoter directs β-glucuronidase expression in Ganoderma lucidum and Pleurotus citrinopileatus. Molecular Biotechnology, 20, 239–244.
Pouresmaeil, M., Dall’Ara, M., Salvato, M. S., Turri, V., & Ratti, C. (2023). Cauliflower mosaic virus: Virus-host interactions and its uses in biotechnology and medicine. Virology. https://doi.org/10.1016/j.virol.2023.02.008
Jiang, P., Zhang, K., Ding, Z., He, Q., Li, W., Zhu, S., Cheng, W., Zhang, K., & Li, K. (2018). Characterization of a strong and constitutive promoter from the Arabidopsis serine carboxypeptidase-like gene AtSCPL30 as a potential tool for crop transgenic breeding. BMC Biotechnology, 18, 1–13.
Bandopadhyay, R., Haque, I., Singh, D., & Mukhopadhyay, K. (2010). Levels and stability of expression of transgenes. Transgenic Crop Plants. https://doi.org/10.1007/978-3-642-04809-8_5
Odell, J. T., Nagy, F., & Chua, N. H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature, 313, 810–812.
Battraw, M. J., & Hall, T. C. (1990). Histochemical analysis of CaMV 35S promoter-β-glucuronidase gene expression in transgenic rice plants. Plant Molecular Biology, 15, 527–538.
Benfey, P. N., Ren, L., & Chua, N. H. (1990). Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO Journal, 9, 1677–1684.
Pratiwi, R. A., & Surya, M. I. (2020). Agrobacterium-mediated transformation. In: Genetic transformation in crops. IntechOpen.
Shou, H., Frame, B. R., Whitham, S. A., & Wang, K. (2004). Assessment of transgenic maize events produced by particle bombardment or Agrobacterium-mediated transformation. Molecular Breeding, 13, 201–208.
Travella, S., Ross, S., Harden, J., Everett, C., Snape, J., & Harwood, W. (2005). A comparison of transgenic barley lines produced by particle bombardment and Agrobacterium-mediated techniques. Plant Cell Reports, 23, 780–789.
Gao, C., Long, D., Lenk, I., & Nielsen, K. K. (2008). Comparative analysis of transgenic tall fescue (Festuca arundinacea Schreb.) plants obtained by Agrobacterium-mediated transformation and particle bombardment. Plant Cell Reports, 27, 1601–1609.
Zhang, Y., Yin, X., Yang, A., Li, G., & Zhang, J. (2005). Stability of inheritance of transgenes in maize (Zea mays L.) lines produced using different transformation methods. Euphytica, 144, 11–22.
Raza, G., Singh, M. B., & Bhalla, P. L. (2019). Somatic embryogenesis and plant regeneration from commercial soybean cultivars. Plants (Basel), 9, 38.
Stolarz, A., Macewicz, J., & Lörz, H. (1991). Direct somatic embryogenesis and plant regeneration from leaf explants of Nicotiana tabacum L. Journal of Plant Physiology, 137, 347–357.
Gill, R., & Saxena, P. K. (1993). Somatic embryogenesis in Nicotiana tabacum L.: Induction by thidiazuron of direct embryo differentiation from cultured leaf discs. Plant Cell Reports, 12, 154–159.
Pathi, K. M., Tula, S., & Tuteja, N. (2013). High frequency regeneration via direct somatic embryogenesis and efficient Agrobacterium-mediated genetic transformation of tobacco. Plant Signaling & Behavior, 8, e24354.
Windels, P., De Buck, S., & Depicker, A. (2008) Agrobacterium tumefaciens-mediated transformation: patterns of T-DNA integration into the host genome. Agrobacterium from biology to biotechnology, 441–481.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by the cooperation of all authors. All authors revised the manuscript critically and also read and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical Approval
Not available.
Consent to Participate
Not available.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Azizi-Dargahlou, S., Pouresmaeil, M. & Ahmadabadi, M. Tobacco Plant: A Novel and Promising Heterologous Bioreactor for the Production of Recombinant Bovine Chymosin. Mol Biotechnol (2024). https://doi.org/10.1007/s12033-023-01043-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-023-01043-z