Abstract
Teak (Tectona grandis Linn. f.) is one of the major plantation timbers of the world. The species is native to India, Myanmar, Thailand and Laos in South East Asia but was translocated to several countries in Africa and Central and South America during the past century. Today, large areas of plantations are grown outside the species native range. It is speculated that genetic bottlenecks and founder effects combined with new selection pressures under new growing conditions have led to the formation of distinct landraces; this hypothesis is supported by results from international provenance tests. In the present study, we apply genetic markers to identify the likely origin of teak grown outside its native range and examine if the landraces show signs of reduced genetic diversity. We find large variation in the level of diversity among landraces, although not larger than that observed among native populations. We conclude that variation in the studied teak landraces probably reflects their areas of genetic origin rather than severe founder effects created during their introduction. The genetic data suggests that the studied landraces originated from either the semi-moist east coast of India, southern Myanmar or western Thailand. These results indicate that translocation of teak has mainly come from a certain part of the native distribution and that this did not include the widespread natural teak areas of southern, dry interior or western India or northern Myanmar.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Teak (Tectona grandis Linn. f.) is an outcrossing deciduous tree of the Lamiaceae family (The International Plant Names Index 2014). It has a large, partly discontinuous natural distribution area encompassing parts of India, Myanmar, Thailand and Laos (Kaosa-ard 1981). Teak is widely known and has an excellent reputation for wood quality, combining strength with lightness, durability, dimensional stability, non-corroding properties, ease of working and seasoning, termite, fungus, chemical, water and weather resistance and attractiveness (FAO 2009). At the current price of US$600–1000 per m3 for high-quality logs and US$350–500 per m3 for low-dimension plantation logs, teak is one of the most expensive major hardwoods in the world (FAO 2015). Today, the species is estimated to be planted in a total of 65 countries outside its native distribution (Koskela et al. 2014).
The history of landrace formation in teak
Teak was probably translocated outside its natural distribution range some 400 to 600 years ago, when it was naturalised in Java, Indonesia (Pandey and Brown 2000; Verhaegen et al. 2010 and references therein). Later plantations were established in Sri Lanka (year 1680) (Perera 1962). In Africa, the Germans introduced teak to Tanzania at the end of the nineteenth century using seed from Calcutta, India (Madoffe and Maghembe 1988). Later, before the First World War, many provenances of teak were introduced by the German colonial administration, with seed sources being from locations such as Tenasserim in southern Myanmar, Travancore in southern India and possibly also from Java (Wood 1967).
In 1902, teak was planted for the first time in Nigeria, with seed first from India and subsequently from Myanmar or Thailand (Chollet 1958; Horne 1966). The species was introduced in Togo and Ghana in 1905 with seed from Nigeria (Chollet 1958) and in the Ivory Coast in 1926 with seed from Togo (Tariel 1966).
In tropical America, the first pure teak plantations were started in 1913 in Trinidad and Tobago (Brooks 1941). This was based on seed from Tenasserim in Myanmar (Beard 1943, here after Keogh 1978), and in the next 3 years, two additional seed lots were imported from the same source (Moore 1966, here after Keogh 1978). This would later become known as the Tenasserim-Trinidad strain (Keogh 1978). Another important introduction in Central America was made in 1926 from Sri Lanka (at that time Ceylon) to Summit Gardens in Panama, where a rather small seed lot produced offspring that was later spread throughout the Panama Canal Zone and surrounding countries. This strain was named the Ceylon-Panama strain (Keogh 1978). Seed of these two above-mentioned strains was widely distributed in the region. A third significant strain, introduced into Honduras from Trinidad in 1927, probably originated in Myanmar, although not necessarily from the same place as the Tenasserim-Trinidad strain (Keogh 1978).
Most of the early introductions mentioned above were on a pilot basis, with large-scale planting only initiated more recently. However, the historical records presented above suggest that later upscaling of plantation forestry was often based on seed from the early introductions. The introduction and domestication history combined with the diverse environmental conditions present in the new teak growing areas could obviously have led to the formation of various landraces. This is supported by observations from international provenance trials where some landraces were found to deviate substantially in growth, stem form or early flowering from populations in the natural distribution area (Keiding et al. 1986). However, it has not been possible to test to what degree the landraces differ from their original source population because the genetic origin of the landraces has been uncertain or imprecisely described in almost all cases.
Present and future supply of teak
By the latest global assessment of teak resources, Kollert and Cherubini (2012) estimated the area of natural teak forests in India, Myanmar, Thailand and Laos to be 29.04 million ha, while a conservative estimate of the teak plantation area was set to 4.35 million ha. The latter presents a considerable increase from the former estimate from the mid-1990s of 2.25 million ha (Pandey and Brown 2000). The teak plantation distribution among regions is also changing. In 1995, about 94% of global teak plantations were in tropical Asia; around 4.8% (109,000 ha) were in tropical Africa, while only 1.5% (34,000 ha) were from tropical America and the Caribbean (Pandey and Brown 2000). Fifteen years later, Kollert and Cherubini (2012) found the African share to constitute 11% of the total (470,000 ha) and the tropical American (Caribbean, Central and South America) share to be 6% (270,000 ha). At the same time, supplies of teak wood from natural forests have started to dwindle since the 1980s, mainly because of logging or log export bans in India, Laos and Thailand (Kollert and Cherubini 2012). Moreover, high-input teak plantations have been developed since the late 1980s, particularly in Latin America. Latin America will thus become increasingly important for plantation supply if the current rate of expansion continues in this region (FAO 2009).
The recent complete map of global teak diversity
Although teak is one of the most important cultivated tropical hardwoods (FAO 2009), studies describing the genetic pattern across the whole natural distribution area by use of DNA markers are sparse. Fofana et al. (2009) made the first attempt and found substantial genetic structure and differences among the native populations. However, this study was based on only 166 individuals from 17 provenances and without any provenances from Myanmar. This latter point is important, since almost half of the remaining natural forests are found in Myanmar (Kollert and Cherubini 2012) and because historical records suggest that Myanmar has been source for several of the first translocations of teak to Africa and America as discussed above. Recently, Hansen et al. (2015) completed the first study of teak’s genetic structure comprising provenances from the total distribution area of the species, including Myanmar. Using simple sequence repeat (SSR) markers on 721 individuals from 29 provenances, a very strong genetic structure was revealed, showing an overall F st value of 0.23 with population pairwise F st values in the range 0.01–0.48. At the same time, large differences in the level of genetic diversity were observed in the different regions. In the eastern part of the distribution area (Myanmar, Thailand and Laos), a strong clinal decrease in diversity was observed towards the east, where the average number of alleles of the southern Myanmar populations was around three times higher compared to the eastern Laos and Thai populations (Hansen et al. 2015). The findings draw attention to the importance of the genetic origin of native as well as planted teak, but the comprehensive study also opens opportunities for the use of so-called assignment methods, which are designed to determine population membership for individuals or groups of individuals using genetic information (Manel et al. 2005). During the last decades, these methods have been increasingly used in wildlife forensic science applications (Ogden and Linacre 2015) including a range of studies that track illegal timber trade of forest tree species (Tnah et al. 2010; Degen et al. 2013).
Objectives of the paper
The objectives of this paper are based on the above observations that
-
Teak has immense importance to global tropical plantation forestry
-
Teak is widely used outside its natural distribution area, which may have led to the formation of distinct landraces often characterised by unknown or imprecisely described genetic origin
-
Future supply of teak timber will increasingly rely on plantations, many in tropical America and Africa
-
Recent DNA technologies and studies have now made it possible to trace the origin of various landraces and compare their level of genetic diversity
The specific objectives of this paper are to
-
1.
Find the genetic origin of major landraces, with special emphasis on tropical America
-
2.
Compare the genetic diversity of the landraces, both among themselves but also to the diversity of their likely origin
For a subset of the landraces tested in field trials, we further wanted to investigate how the landraces perform compared to populations of native origin.
Materials and methods
Plant material—landraces
Six populations from Africa, seven from Latin America and four from Indonesia were included in the study. These 17 populations were obtained from various sources, which in the rest of this paper are collectively designated as landraces. Four were sampled as leaf material from a provenance trial in Longuza, Tanzania (Persson 1971; sampling location labelled ‘Longuza’ in Table 1). One represents a commercial seed lot from Tanzania (sampling location labelled ‘Tanzania’ in Table 1). Four populations, sampling location labelled ‘FIP38’ in Table 1, were collected from the provenance trial at Pha Nok Kao in Thailand (trial no. 038), a part of the international series of teak provenance trials described by Keiding et al. (1986). Sampling of trees in the provenance trials for the present study was done without any selection (consecutive remaining trees up to 31 individuals). An additional six populations were shipped as seeds from commercial teak planting companies, which are members of Camcore, an international tree breeding and conservation programme with headquarters at North Carolina State University (http://www.camcore.org; sampling location labelled ‘Camcore’ in Table 1). Seeds were collected separately from 30 (or 15) trees and germinated, and one seedling from each tree-seed lot was used for genotyping. Finally, two populations were collected as leaf material from two commercial plantations in Panama (sampling location labelled ‘IWC’ in Table 1).
Plant material—genetic reference populations
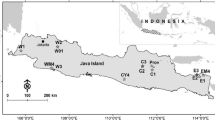
The 29 teak populations originating from the natural distribution area and analysed by Hansen et al. (2015) using six SSR markers were used as a reference population to constitute a ‘genetic map’ to which we could try to assign the landraces described above. Further details about the origin and sampling of the 29 populations are given in Hansen et al. (2015) and references therein. Bayesian cluster analysis of the 29 populations analysed by Hansen et al. (2015) resulted in three main clusters and within those, six sub-clusters— the latter are depicted together with the geographic origin of the 29 populations in Fig. 1.
Map showing the origin of the 29 natural populations of teak which constitute the reference population in the current study. The six sub-clusters identified via cluster analysis by Hansen et al. (2015) are depicted by colour coding. The dotted line indicates the approximate outer boundaries of the natural distribution of teak
Laboratory work
DNA from the landraces was extracted from the leaf tissue using any one of three methods: (1) the QIAGEN® (Germany) DNeasy Plant Mini Kit, (2) the QIAGEN® DNeasy 96 Plant Kit or (3) a modified version of the CTAB method of Doyle and Doyle (1990). All individuals were genotyped with six microsatellites developed by Verhaegen et al. (2005): CIRAD1TeakA06, CIRAD1TeakB03, CIRAD2TeakB07, CIRAD2TeakC03, CIRAD3TeakA11 and CIRAD3TeakF01.
Genotyping of microsatellites took place in 10-μl PCR reactions using the QIAGEN® Multiplex kit (catalogue no. 206143). PCR conditions followed the given standard multiplex PCR protocol: 1× Multiplex master mix (providing a final concentration of 3 mM MgCl2), 0.2 μM of each primer and around 20 ng of DNA sample with water added to make the final reaction volume. Amplifications were carried out in a Bio-Rad thermal cycler (model C1000) with the following thermal profile: 15 min of denaturation at 95 °C, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 90 s and extension at 72 °C for 60 s, with a final extension step at 60 °C for 30 min. Fragment sizes were determined on an ABI 3130XL genetic analyser and analysed with the GeneMapper software version 4.0 (Applied Biosystems). Genotyping of both genetic reference populations and landraces was done by the same methods on the same equipment.
Genetic diversity and structure
Genetic diversity in the landraces was estimated with the following parameters: observed heterozygosity (H o ), expected heterozygosity (H e ), number of observed alleles (Na), effective number of alleles (Ne) and allelic richness calculated via rarefaction (Na(rar)). The first four parameters were calculated in GenAlEx vers. 6.5 (Peakall and Smouse 2006, 2012), and allelic richness was calculated in HP-Rare vers. 1.1 (Kalinowski 2005). Na is the average number of observed alleles while Ne estimates the number of equally frequent alleles that it would take to achieve a given level of genetic diversity in a panmictic population (usually less than the actual number; Kimura and Crow 1964). Na(rar) is the number of observed alleles corrected for the effects of sample size through rarefaction, a correction that is important since larger sample sizes are expected to sample more alleles than smaller samples (Kalinowski 2004). The fixation index, F is (Hartl and Clark 1997), was also calculated in GenAlEx.
To explore the genetic differentiation among the landraces, pairwise F st values between populations were calculated in GenAlEx based on the framework of an analysis of molecular variance (AMOVA; Excoffier et al. 1992). In addition to this traditional measure of genetic differentiation, G″ st values (Meirmans and Hedrick 2011) were calculated and tested with G-statistics in GenAlEx as an alternative estimator for population differentiation. The latter was developed specifically to take the high polymorphism of microsatellites into account, which otherwise may result in underestimation of the genetic differentiation (Hedrick 2005).
As a means of graphically illustrating the calculated genetic differentiation among landraces, while at the same time showing how differentiated the self-same landraces are to natural populations from different regions of the natural distribution, pairwise G″ st values were also calculated between all natural populations in the reference population and all 17 landraces. The resulting G″ st value matrix with 46 columns/rows was used as input to construct a neighbour-joining (NJ) tree (Saitou and Nei 1987); this was done in the TreeFit software (Kalinowski 2009), which also calculates the proportion of variance in the genetic distance matrix that is explained by the tree (R 2). The resultant tree was depicted in the graphical viewer Dendroscope 3 ver. 3.5.7 (Huson and Scornavacca 2012).
In an attempt to explore the origin of the 17 landraces, a Bayesian cluster analysis was conducted using the STRUCTURE software (version 2.3.4) (Pritchard et al. 2000), whereby all genetic data from the populations originating from both the natural distribution and from the landraces was included. The USEPOPINFO model was used, whereby the population origins of the 29 natural populations were used to estimate the ancestry of populations of unknown origin (the 17 landraces). Technically, this option is switched on in the front end using the option “Update allele frequencies using only individuals with POPFLAG = 1”, located under the “Advanced Tab” (Pritchard et al. 2010) and where learning samples are implemented using the PopFlag column in the data file. The predefined population (the natural population) was used for those individuals for whom PopFlag = 1 (and whose PopData is in (1…K)). The PopData value is ignored for individuals for whom PopFlag = 0. Ancestry of individuals with PopFlag = 0 is updated according to the admixture or no-admixture model, as specified by the user (Pritchard et al. 2010). As in the analyses of natural populations by Hansen et al. (2015), two rounds of STRUCTURE analyses were run. The first one comprised data from all 46 populations (1166 trees) and used the ‘no admixture ancestry’ model in combination with the assumption of independent allele frequencies among populations, due to the strong genetic structure observed in the overall natural distribution area. In the second round of STRUCTURE analyses, separate analyses were run on data sets comprising the populations from the main clusters resulting from the first round of STRUCTURE analysis. In the second round, the ‘admixture ancestry’ model combined with the assumption of allele frequencies being correlated among populations was applied, as the natural populations within the main clusters are geographically closer. In both rounds, ten clustering runs were made for each K (from 1 to 8 in round 1; from 1 to 5 in round 2), each with a burn-in time and run length of 100,000. To infer the true number of clusters (K), we used the delta K method developed by Evanno et al. (2005) and the STRUCTURE HARVESTER software (Earl and vonHoldt 2012) to implement the method. As clustering in STRUCTURE involves stochastic simulation, replicate cluster analysis of the same data may produce different solutions, due either to ‘label switching’ or ‘genuine multimodality’ (Jakobsson and Rosenberg 2007). Consequently, we used the CLUMPP software (Jakobsson and Rosenberg 2007) to align the outcome of the ten replicate cluster analyses with the identified optimal number of clusters (K). The DISTRUCT software (Rosenberg 2004) was used to illustrate the outcome of this alignment.
Genetic assignment
Another approach to unravel the genetic origin of individuals or populations is to apply assignment tests, which are methods designed to determine population membership for individuals (Guinand et al. 2002) based on multilocus genetic data, using both individual genotypes and population-level allele frequencies (Davies et al. 1999). Using the genetic data from the study of teak populations covering the full range of the natural distribution (Hansen et al. 2015) as an overall reference population of 721 individuals, the 445 individuals from the 17 landraces were assigned to their most likely origin. This was done in the software GENECLASS2 (Piry et al. 2004), and two different assignment methods were applied: a frequency method developed by Paetkau et al. (1995) and a Bayesian method developed by Rannala and Mountain (1997). Both methods included simulation of 1,000,000 individuals. We tested the assignment power by performing a self-assignment: all trees from the overall reference population (= known origin) were assigned to the most likely reference (sub)population according to their genotype. The individual to be assigned was excluded from its respective (sub)population during computations of assignment criteria (leave-one-out procedure; Efron 1983) to ensure cross-validation. An assignment score for each individual was calculated based on the probability of that individual belonging to the correct population, divided by the sum of probabilities for the individual belonging to each of the potential source populations. The mean value of the scores from all individuals constitutes a so-called ‘quality index’ for the assignment procedure (Piry et al. 2004). Based on the percentage of correctly self-assigned individuals and the quality index of the self-assignment study, we evaluated the scale to determine to which level the procedure allowed reliable assignment: clusters, sub-clusters or populations.
Genetic relatedness and inbreeding within landraces and natural populations
In order to investigate relatedness and inbreeding between individuals within the landraces from the three regions (Latin America, Africa and Indonesia) and compare with the relatedness and inbreeding in the natural populations from their most likely origin, analyses were performed in the software COANCESTRY version 1.0.1.5 (Wang 2011). The two landraces Mozambique (MOZ) and 3047 were omitted from these analyses since they did not cluster together with the other landraces from Africa and Indonesia, respectively. Several relatedness estimators have been proposed, but no single estimator performs best in all situations as the rank order of these estimators is marker data dependent and relies on the true relatedness being estimated or on the population’s relatedness structure (Wang 2011). Consequently, COANCESTRY offers the possibility to make a simulation using the available marker data and test efficiency of seven different estimators based on simulated individuals of known relationship (e.g. half-sibs). We did this with the SSR data for the sub-cluster consisting of natural populations 3033, 3034 and 3039 and the four populations from southern Myanmar, since most landraces appeared to originate from this region. Based on the simulations, we selected three relatedness estimators: the triadic likelihood estimator by Wang (2007) (TrioML), the moment estimator described in Ritland (1996) (Ritland) and the moment estimator given in Queller and Goodnight (1989) (QuellerGt). For the same pairs of groups, inbreeding coefficient (F) point estimates from the Ritland estimator and from a moment estimator described by Lynch and Ritland (1999) were calculated. The difference in both average relatedness and inbreeding coefficient between groups of natural populations and landraces was tested by a permutation procedure in COANCESTRY, using 100,000 bootstraps.
Phenotypic data
We reanalysed published data from the series of international provenance trials (Keiding et al. 1986): trials FIP038 (Thailand) and FIP032 (Papua New Guinea) to compare the performance of the landraces from Indonesia with populations from the natural distribution, trial FIP022 (Mexico) to compare the performance of the landraces from Latin America with populations from the natural distribution and trial FIP008 (Ghana) to compare the performance of West African landraces with populations from the natural distribution. We further used published results from Madoffe and Maghembe (1988) to compare the performance of East African landraces with natural populations in the Longuza trial (Tanzania). We included data on average growth rate (basal area-weighted mean diameter) and persistence of main stem axis (related to stem form) assessed at age nine from the FIP trials (details in Keiding et al. 1986) and data on growth rate (basal area per ha) and stem straightness in the Tanzanian trial (details in Madoffe and Maghembe 1988). Both axis persistence and stem straightness were scored on scales where higher values correspond to better stem quality. All trials were established as replicated block trials, where significant differences were found between the provenances. We compared the performance of the landraces with natural populations based on their mean performance. Four of the provenances in the Mexican FIP022 trial had survival rates below 40% and were therefore excluded from the comparison.
Results
Genetic diversity and differentiation of landraces
Substantial genetic differences among both regions and landraces were observed for the diversity parameters (Table 2). Despite variation in sample size (range of N = 13–35), the observed number of alleles (Na) and the allelic richness calculated via rarefaction (Na(rar)) showed a Pearson’s correlation of 0.97 (p < 0.001); therefore, Na represents a suitable measure for comparison of diversity among the studied populations.
The four landraces from Indonesia were rather similar: all had a genetic diversity below average (range of Na(rar) 4.12–4.87). Also, the seven landraces from Latin America and the Caribbean constituted a rather homogenous group with regard to genetic diversity (range of Na(rar) 5.33 (TRI_Har)–6.56 (COL)). One landrace stuck out as extremely low in genetic diversity, namely the one from MOZ, having an expected heterozygosity (H e ) of only 0.24 and an average of 1.83 alleles per loci (Na) (compared to overall means of 0.65 and 6.11 for these two parameters, respectively). The two landraces that showed the highest diversity were also from Africa, namely TAN_Mti from Tanzania and IVO from the Ivory Coast (Table 2).
The majority of the studied landraces were estimated to have a fixation index (F is ) close to zero (overall average = 0.01), with a few moderate deviations from zero (TAN_Lon = 0.13, MOZ = 0.08 and VEN and 3050 = 0.07). The highly significant AMOVA gave an overall F st value of 0.126 (among population variance), while population pairwise F st values were in the range 0.00 to 0.46 (Table 3—lower triangle). Applying G-statistics and the G″ st differentiation parameter developed by Meirmans and Hedrick (2011), the overall differentiation was estimated to be 0.362. The pairwise G″ st values were in the range 0.00 to 0.82 (values in upper triangle of Table 3). The Pearson’s correlation between pairwise estimates of F st and G″ st was 0.94 (p < 0.001). The most divergent landrace was MOZ, which also contained very low levels of diversity. The smallest pairwise F st value for MOZ was 0.24 shared with CAM_INDO, and in general, MOZ was most similar to the landraces from Indonesia (range 0.24–0.33) and Tanzania (range 0.31–0.35). Among the remaining African landraces, we found TAN_Mti to be quite differentiated from the other two Tanzanian landraces. This fits well with the historical records of early import to East Africa from various sources spanning from Myanmar, Java and/or Southern India. TAN_Kih and TAN_Lon are reported to have the same genetic origin, while TAN_Mti is likely to have a different genetic origin (Table 1). After the African landraces, the four Indonesian landraces showed the largest within-region population differentiation (range 0.02–0.09 for pairwise F st values) although the group still presents a relatively uniform cluster compared to the overall genetic differentiation (F st = 0.126). The tested Latin American and Caribbean landraces represented a very uniform group (range 0.00–0.06 for pairwise F st values) with several pairwise F st values not statistically significant different from 0 (colour coding in Table 3).
The above results are reflected in the NJ tree in Fig. 2. The tree explained 76% of the variance in the matrix of pairwise G″ st values among the total of 46 available populations (landraces + natural populations). All of the seven landraces from Latin America and the Caribbean are clustered closely together in the lower left corner, together with populations from southern Myanmar. The landraces from Africa are scattered between Thai and Lao origins but have no close relation to any specific natural population from the region. Three of the four Indonesian landraces are positioned fairly closely within the tree. The Indonesian population 3047 seems to be more closely related to some western Thai populations, yet the Indonesian landraces are obviously closer to the Thai/Laotian group compared to India or Myanmar. None of the studied landraces were found to be closely related to either populations from the dry interior of India, the moist west coast of India or northern Myanmar: these regions constitute distinct groups in the lower branches of the tree (Fig. 2).
Neighbour-joining tree using the pairwise population matrix of G″ st values as input (genetic distance matrix). Populations from the natural distribution area are coloured as done in Hansen et al. (2015). Colour code: yellow = India—dry interior, dark green = India—moist west coast, magenta = India—semi moist east coast, light green = Myanmar—northern region, cyan = Myanmar—southern region, blue = Thailand, red = Laos, black = landraces
Clustering of landraces among native populations
The findings based on pairwise genetic differences were supported by the Bayesian cluster analysis using STRUCTURE and comprising both the 17 landraces and the 29 natural populations. The STRUCTURE analysis revealed that the most likely number of clusters (K) was three, using the delta K method (Evanno et al. 2005). Six of the 17 landraces, all from Latin America and the Caribbean, distributed to cluster 2 (orange), encompassing the natural populations from the semi-moist east coast of India and southern Myanmar; seven landraces are admixtures of clusters 2 and 3 (blue) while the remaining four landraces (three from Indonesia and the one from Mozambique) predominantly belonged to cluster 3 (blue) comprising the natural populations from Thailand and Laos (Fig. 3). Only three landraces from Africa (TAN_Lon, NIG and IVO) and one from Latin America (COL) showed small indications of having part ancestry in cluster 1 (light green), consisting of Indian populations from the dry interior and the moist west coast, although predominantly all of these landraces seem to originate from cluster 2 (Fig. 3).
Results from STRUCTURE analysis with K = 3 clusters, where the ten replicate cluster analyses have been aligned in CLUMPP. Each of 46 populations (1166 trees in total) is represented with a horizontal bar. Vertical height of the bar represents the number of trees in the population (range 10–35). The first 29 populations (from above) are natural populations—the last 17 are landraces. For each population, an estimated cluster membership coefficient is shown; bar colours indicate the clusters: (1) light green = India—dry interior and moist west coast, (2) orange = India—semi-moist east coast and Myanmar and (3) blue = Thailand and Laos
In the second round of STRUCTURE analyses, separate analyses were run on the populations from the main clusters resulting from the first round, though no second-round analyses were conducted for cluster 1, since no landraces clustered with these populations. The two main clusters (clusters 2 and 3) used as starting points had slightly different compositions with regard to natural populations than clusters 2 and 3 from Hansen et al. 2015 (see Fig. 1), since we here (correctly) assigned population 3036 from eastern India to cluster 1 (and not cluster 2) and also (correctly) assigned population 3039 from western Thailand to cluster 2 (and not cluster 3) (compare Figs. 1 and 3). Two sub-clusters were revealed within each of the main clusters (2 and 3) using the delta K method. These four sub-clusters are depicted in Fig. 4. The main cluster 2 (from the first STRUCTURE analysis) separated into one sub-cluster consisting of the four northern Myanmar populations (coloured light blue) and a second sub-cluster (coloured brown) comprising of two populations from eastern India, one from west Thailand, four from southern Myanmar and 13 landraces (Fig. 4a). Figure 4b illustrates the two sub-clusters from main cluster 3, where the first sub-cluster (coloured purple) is made up by the most eastern of the natural Thai populations plus the two Laotian populations, while the second sub-cluster (coloured light pink/white) comprises two natural populations more to the west in Thailand (3041 and 3042) and the four Indonesian landraces. The Mozambique landrace also fell into this eastern cluster. The results from the STRUCTURE analyses were thus in good agreement with the NJ tree based on G″ st values (Fig. 2).
Results from second round of STRUCTURE analyses. Since no landraces were clustered together with the populations from the dry interior and moist west coast of India (main cluster 1), no second round of analysis was made for these populations. Within the two remaining main clusters, two sub-clusters were identified in each. Graphs a and b show ten replicate cluster analyses for K = 2 which have been aligned in CLUMPP
Assignment of landraces to likely population of origin
The fraction of correctly self-assigned individuals was 91.7% (661 individuals) when the reference population was split up into three main STRUCTURE clusters using the Bayesian assignment method, while the corresponding result was 53.7% (387 individuals) when individuals were self-assigned to the 29 individual populations (for the total comparison of results from self-assignment, see Table 4). However, both correct self-assignment and quality index were acceptable when based on the level of six sub-clusters (Table 4) and this scale also makes sense given the density of the sampling of reference populations from the native range. We therefore decided to assign the 17 landraces at this level, and results from this assignment are presented in Table 5. The two different assignment methods gave very similar results; therefore, below, we only refer to the results of the Rannala and Mountain (1997) method.
All the African landraces, apart from the odd Mozambique population, had around 90% or more of their individuals assigned to the sub-cluster consisting of natural populations from north-eastern India, west Thailand and southern Myanmar (C2_SC1). For the landraces from Latin American and the Caribbean, the picture was even clearer; in all seven landraces, 100% of the individuals were assigned to this sub-cluster.
The four Indonesian landraces showed a mixed pattern, with 87% of the individuals in 3047 assigned to the eastern India, west Thailand and southern Myanmar (C2_SC1), while individuals from 3049 were assigned exactly 50:50 between C2_SC1 and C3_SC2, the latter consisting of two natural Thai populations (3041 and 3042). For 3050, the majority of individuals (68%) were assigned to C3_SC2, while for CAM_INDO, 57% were assigned to C2_SC1. Overall, there is a high degree of correspondence between the assignment tests and the STRUCTURE results. The only landrace that seems to provide a slightly different result between these two approaches is CAM_INDO, which in STRUCTURE grouped predominantly with the main cluster 3 (Thailand and Laos), while in assignment tests was placed in the southern Myanmar and east Indian sub-cluster C2_SC1.
Levels of relatedness within populations
Results from the comparative analysis of within-population relatedness and inbreeding coefficients between landraces from the three regions (Latin America, Africa and Indonesia) and the natural populations from the distribution area where the landraces were assigned are seen in Fig. 5.
Results from comparative analysis of within-population relatedness and inbreeding coefficient between landraces and the natural populations from the distribution area where the landraces were assigned. Upper three panels: comparison of relatedness among individuals within populations. Lower three panels: comparison of inbreeding coefficient within populations. The difference in relatedness and inbreeding coefficient between natural populations and landraces was tested by 100,000 bootstraps
In the upper three panels of the figure, a comparison of relatedness within natural populations with relatedness within landraces from the three non-indigenous regions is presented. In the lower three panels, the inbreeding coefficients are compared for the same populations and landraces. For the comparison of landraces from Latin America and the Caribbean and from Africa with natural populations from sub-cluster C2_SC1 (southern Myanmar and east India), which is the natural distribution area where they all seem to originate, there is a statistically significant trend of higher relatedness within the landraces for all three estimators. The only exception is for the QuellerGt estimator in the Africa comparison; however, this is non-significant. In the case of the Indonesian landraces (rightmost upper panel in Fig. 5), the trend seems opposite, with statistically higher relatedness in the natural populations for two of the estimators, while the difference was non-significant for the last estimator (TrioML). However, this comparison is based on only two natural populations in the sub-cluster C3_SC2 (3041 and 3042) and only three landraces (3049, 3050 and CAM_INDO).
In the comparison of the inbreeding coefficient (F) (lower three panels in Fig. 5), the trend is a higher F in the natural populations compared with the landraces from all three regions (except for LynchRD in the Africa comparison). All the differences are non-significant but are based on a much lower number of data points (individuals’ F vs. thousands of dyads).
Performance of the landraces
The results from the five trials consulted in the present study are presented in Fig. 6 where performance in relation to stem form is plotted against growth data. In the Mexican trial, large variation was observed among the Latin American landraces in both growth and stem quality, with landraces presenting both good- and poor-performing provenances compared with the natural populations (Fig. 6a). The Indonesian/Papua New Guinean landraces tested in the Thai and Papua New Guinean trials were among the fastest-growing landraces but displayed average or below average stem quality (Fig. 6b, c). The local landraces tested in the trial in Ghana were among the slowest-growing landraces and particularly had lower stem quality compared with provenances from the natural populations (Fig. 6d). Still, the Indonesian landraces were among the fastest-growing landraces in this trial. Finally, the east African landraces in the Tanzanian trial were fast growing with good quality compared with the natural populations, although a single provenance from moist west India (Coimbatore) was the best performing (Fig. 6e).
Variation in average stem form and growth rate among various landraces and natural origins in five provenance trials. a Mexico, IP022. b Papua New Guinea (PNG), IP032. c Thailand, IP038. d Ghana, IP008. e Tanzania, Longuza. High values of persistence/straightness refer to superior stem form. Based on data from Keiding et al. (1986) (a–d) and Madoffe and Maghembe (1988) (e)
Discussion and conclusions
In the present study, we used the first ‘global genetic reference map’ made by Hansen et al. (2015) to infer the likely genetic origin of major landraces, with special emphasis on tropical America, and we compared the genetic diversity among landraces to the diversity levels present in natural populations from their likely origin.
We found that of 17 investigated landraces from Indonesia, Africa and tropical America, the majority most likely originated from southern Myanmar or semi-moist east coast of India; the genetic analysis suggests only some of the Indonesian landraces to have a likely origin further east in Thailand.
These findings fit well with the historical records of Tenasserim (southern Myanmar) and Java as likely origins to East African landraces. The Tanzanian Kihuhwi source (TAN_Kih) is genetically very similar to the western Java (3047) plantation, while the Mtibwa plantation (TAN_Mti) is more genetically similar to the native Pyinmana teak forest (MY_PMUA; approximately 300 km north of Yangon, but still in the Southern Myanmar cluster) (Fig. 2). However, we find no signs of the Travancore (southern India) origin mentioned in Wood (1967) as a potential source for Tanzanian landraces.
The seven landraces from Central America were genetically quite similar to each other and to the native Letpadan teak forest (MY_LUA; approximately 120 km north-west of Yangon) (Fig. 2). The results support that they originate from a limited number of introductions of teak into Central America in the beginning of the twentieth century with Myanmar as an important source (both a Tenasserim strain and an additional import from Myanmar) as suggested by Keogh (1978). Keogh (1978) also highlights a potential important introduction to Central America (through Panama) based on an original collection from trees in a botanical garden in Sri Lanka. We have no knowledge of the genetic origin of the trees in the botanical garden, but teak is not native to Sri Lanka and one can therefore speculate that the southern Indian teak forest has been the likely source due to geographic proximity to Sri Lanka. However, our genetic study reveals no signs of the south Indian cluster in the analysed Central American landraces. We cannot exclude that the studied Central American plantings originate from this route, because trees in the Sri Lanka botanical garden could have been grown from seed collected from trees growing somewhere in the east India-south Myanmar cluster. The most likely origin of the two West African plantations in the present study was also the east India-south Myanmar cluster, where the sample from Nigeria clustered out with the Central American landraces, while the Ivory coast seemed more similar to the Tanzanian landraces (Fig. 2). Interestingly, none of the studied landraces seem to have originated from the natural distribution of teak in southern or western India or from northern Myanmar.
A few other studies have dealt with the genetic origin of teak landraces. The findings of the present study are generally in concordance with results from Verhaegen et al. (2010). Their analysis supported that the African teak mainly originates from northern India, which is not contradictory to our findings, but they had fewer natural reference populations and no populations from Myanmar. Verhaegen et al. (2010) further deduced that the Indonesian teak, of which three landraces are the same as in our study (3047, 3049 and 3050), is closely linked to central Laos or eastern Thailand. Our results place the origin of Indonesian teak further west in north-western Thailand. One Indonesian landrace (3047) is even assigned to the north Indian/south Myanmar sub-cluster. Our analysis assigned 13 landraces to the sub-cluster composed of two east-Indian, four south-Myanmar and one west-Thai populations, but the analysis could not reveal if they were most likely to originate from India or Myanmar/western Thailand. In a similar way, Huang et al. (2015) used a provenance trial in south China to try to determine the genetic relationships between 18 natural populations from four different countries and ten introduced provenances, of which the majority of the latter had been planted in China without any record of their origins. Their analyses revealed that the introduced provenances had probable origins in Laos, Thailand or Myanmar, while there was no sign of a genetic relationship with the four natural populations from India that were included in the study. The issue regarding the genetic origin of teak takes on a new dimension when plantations are established within the natural range of the species. Recently, DNA marker studies of teak in Myanmar (Minn et al. 2014; Thwe-Thwe-Win et al. 2015) revealed substantial variation in genetic diversity and genetic structure within the large and extensive teak area in the country. Teak planting in Myanmar was initiated in the 1700s, but until 2007, it was conducted by governmental institutions using native seed from Myanmar (Thwe-Thwe-Win et al. 2016). Since 2007, private companies have established large-scale teak plantations in Myanmar using both native seed and seed introduced from Indonesia and China, for example. Thwe-Thwe-Win et al. (2016) compared the genetic diversity of some of these young plantations with the natural populations and found that the genetic composition of plantations was often substantially different from the nearby natural populations. They further found that some of the plantations based on exotic seed had markedly lower genetic diversity than the native populations. Both phenomena may pose a potential threat to the genetic diversity and structure that constitute the genetic resource of teak in Myanmar (Thwe-Thwe-Win et al. 2016).
The overall genetic differentiation among the 17 landraces (F st value of 0.126) in the present study is considerably smaller than the overall F st value of 0.227 among natural populations found by Hansen et al. (2015) for the entire distribution area. The result reflects that even though landraces are found on different continents, they mainly originate from a certain part of teak’s natural distribution area, as revealed in the present study from the NJ tree (based on pairwise G″ st values), STRUCTURE analysis and assignment tests. According to our analysis, none of the 17 landraces were thus related to populations from the dry interior of India, the moist west coast of India or northern Myanmar.
Various international provenance trials have shown that a number of economically important quality parameters of plantation grown teak timber depend on the genetic origin: growth rate, stem form, branch size, epicormic branching, amount of heartwood, wood quality, buttressing, survival and health (Kjær et al. 1995; Kjær et al. 1999; Goh et al. 2013a). It is therefore important to understand the genetic base available for planting programmes. Our study, in combination with previous studies (Verhaegen et al. 2010; Huang et al. 2015; Thwe-Thwe-Win et al. 2015), seems to indicate that Indian teak is not widely used outside India.
Differences in genetic diversity among landraces were observed, but only one landrace from Africa (Mozambique) showed very low genetic diversity, indicating a major bottleneck. The seed collection in Mozambique was conducted at a small plantation with low stocking, where a distance of 100 m between selected trees could not be maintained. The possibility of related mating within the small population of unknown origin is a distinct possibility. Overall, the findings suggest that variation in diversity levels among teak landraces in general reflects their relative areas of genetic origin rather than severe founder effects created during their introductions from Asia to Africa and the Americas.
The average diversity level H e = 0.64 and N a = 6.7 for the seven Central America landraces in our study is only slightly smaller compared with the levels H e = 0.68 and N a = 7.5 found for the natural population in southern Myanmar (Table 3 in Hansen et al. 2015), which are the areas we here identify as the likely origin of these landraces. As another example, the three Tanzanian landraces actually had a higher diversity level (average of H e = 0.70 and N a = 6.3) compared to the average level for natural populations in Thailand (H e = 0.52 and N a = 4.3) and Laos (H e = 0.35 and N a = 2.6) reported by Hansen et al. 2015.
The comparisons of within-population relatedness and inbreeding between landraces and natural populations from the putative geographic origin of the very same landraces gave no clear results. This may be due to the fact that it is questionable whether the clusters from the genetic STRUCTURE analysis of the natural populations can be regarded as true reference populations. However, there was a tendency that some of the landraces—especially the ones from Latin America—had a higher relatedness – which theoretically fits well with the historical records claiming that only few introductions of teak have been made to this part of the world. On the contrary, the overall tendency—although not statistically significant—was that the inbreeding coefficient was higher within the natural populations compared to the landraces.
Large potential exists for the improvement of timber quality through breeding (Kjær and Foster 1996; Goh et al. 2013b), but it is important to build breeding programmes on genetically healthy and diverse plant material. It is therefore an important finding that the major landraces, in general, seem to be genetically diverse and not to have suffered from severe bottlenecks during their translocation. The international provenance trials tend to support that major landraces often represent healthy and viable seed sources: our survey of results from the five trials showed that the landraces were generally fast growing and have good survival rates, despite some of the west African landraces revealing low stem quality. Still, single plantations or even planting programmes may originate from seed collected on few trees and negative selection for early flowering could easily have occurred at earlier stages. It is therefore important to mobilise a suitable and diverse founder population at the national or regional level when initiating domestication programmes.
The present study only represents a small sample of the landraces that may have developed in the many teak-growing regions worldwide. However, we trust that the findings have contributed new important information to the general understanding of the origin of landraces in teak. Also, we hope that the results from the study can be used in future more detailed studies of the diversity, genetic quality and dynamics of domesticated teak outside its natural distribution area.
References
Beard JS (1943) The importance of race in teak, Tectona grandis L. Caribb Forester 4(3):135–139
Brooks RL (1941) Notes on pure teak plantations in Trinidad. Caribb Forester 3(1):25–28
Chollet MA (1958) Le teck au Togo. Bois For Trop 49:9–18
Davies N, Villablanca FX, Roderick GK (1999) Determining the source of individuals: multilocus genotyping in nonequilibrium population genetics. Trends Ecol Evol 14:17–21
Degen B, Ward SE, Lemes MR, Navarro C, Cavers S, Sebbenn AM (2013) Verifying the geographic origin of mahogany (Swietenia macrophylla King) with DNA-fingerprints. Forensic Sci Int Genet 7:55–62
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Efron B (1983) Estimating the error rate of a prediction rule—improvement on cross-validation. J Am Stat Assoc 78:316–331
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics 131:479–491
FAO (2009) The future of teak and the high-grade tropical hardwood sector. Planted Forests and Trees Working Paper FP/44E. Accessed on http://www.fao.org/docrep/012/k6549e/k6549e00.pdf
FAO (2015) Global teak trade in the aftermath of Myanmar’s log export ban by Kollert W & Walotek PJ Planted forests and trees working paper FP/49/E. Rome, Italy (available at http://www.fao.org/forestry/plantedforests/67508@170537/en/)
Fofana IJ, Ofori D, Poitel M, Verhaegen D (2009) Diversity and genetic structure of teak (Tectona grandis L.f) in its natural range using DNA microsatellite markers. New For 37:175–195
Goh DKS, Bacilieri R, Chaix G, Monteuuis O (2013a) Growth variations and heritabilities of teak CSO-derived families and provenances planted in two humid tropical sites. Tree Genet Genomes 9(5):1329–1341
Goh DKS, Japarudin Y, Alwi A, Lapammu M, Flori A, Monteuuis O (2013b) Growth differences and genetic parameter estimates of 15 teak (Tectona grandis L.f.) genotypes of various ages clonally propagated by microcuttings and planted under humid tropical conditions. Silvae Genet 62:196–206
Guinand B, Topchy A, Page KS, Burnham-Curtis MK, Punch WF, Scribner KT (2002) Comparisons of likelihood and machine learning methods of individual classification. J Hered 93:260–269
Hansen OK, Changtragoon S, Ponoy B, Kjær ED, Minn Y, Finkeldey R, Nielsen KB, Graudal L (2015) Genetic resources of teak (Tectona grandis Linn. f.)—strong genetic structure among natural populations. Tree Genet Genomes 11:802
Hartl DL, Clark AG (1997) Principles of population genetics, 3rd edn. Sinauer Associates, Inc., Sunderland
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Horne JEM (1966) Teak in Nigeria. Nigerian information bulletin (New Series) No. 16
Huang GH, Liang KN, Zhou ZZ, Xu JM, Ma HM (2015) Genetic variation and origin of teak (Tectona grandis L.f.) native and introduced provenances. Silvae Genet 64:33–46
Huson DH, Scornavacca C (2012) Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol 61:1061–1067
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Kalinowski ST (2004) Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv Genet 5:539–543
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189
Kalinowski ST (2009) How well do evolutionary trees describe genetic relationships among populations? Heredity 102:506–513
Kaosa-ard A (1981) Teak (Tectona grandis L.f.)—its natural distribution and related factors. Nat Hist Bull Siam Soc 29:55–74
Keiding H, Wellendorf H, Lauridsen EB (1986) Evaluation of an international series of teak provenance trials. Danida Forest Seed Centre, Humlebæk
Keogh RM (1978) Teak (Tectona grandis) provenances of the Caribbean; Central America; Venezuela and Colombia. Unpublished. Original document at FAO headquarters, Rome. 16 p
Kimura M, Crow JF (1964) Number of alleles that can be maintained in finite population. Genetics 49:725–738
Kjær ED, Foster GS (1996) The economics of tree improvement of teak (Tectona grandis L.).Technical Note no 43. The World Bank/Danida Forest Seed Centre. DK-3050 Humlebæk. Denmark. ISSN 0902–3224, p. 22
Kjær ED, Lauridsen EB, Wellendorf H (1995) Second evaluation of an international series of teak provenance trials. Danida Forest Seed Centre, Humlebæk 117p
Kjær ED, Kajornsrichon S, Lauridsen EB (1999) Heartwood, calcium and silica content in five provenances of teak (Tectona grandis L.f. ). Silvae Genet 48:1–3
Kollert W, Cherubini L (2012) Teak resources and market assessment 2010. FAO Planted Forests and Trees Working Paper FP/47/E, Rome. Available at http://www.fao.org/3/a-an537e.pdf. (Accessed February 1 2016)
Koskela J, Vinceti B, Dvorak W, Bush D, Dawson IK, Loo J, Kjaer ED, Navarro C, Padolina C, Bordács S, Bordacs S, Jamnadass R, Graudal L, Ramamonjisoa L (2014) Utilization and transfer of forest genetic resources: a global review. For Ecol Manag 333:22–34
Lynch M, Ritland K (1999) Estimation of pairwise relatedness with molecular markers. Genetics 152:1753–1766
Madoffe SS, Maghembe JA (1988) Performance of teak (Tectona grandis L.f.) provenances seventeen years after planting at Longuza, Tanzania. Silvae Genet 37:175–178
Manel S, Gaggiotti OE, Waples RS (2005) Assignment methods: matching biological questions with appropriate techniques. Trends Ecol Evol 20:136–142
Meirmans PG, Hedrick PW (2011) Assessing population structure: F(ST) and related measures. Mol Ecol Resour 11:5–18
Minn Y, Prinz K, Finkeldey R (2014) Genetic variation of teak (Tectona grandis Linn. f.) in Myanmar revealed by microsatellites. Tree Genet Genomes 11:802
Moore D (1966) The formation of teak plantations by the Group Planting System. Proceedings of the 6th World Forestry Congress, Vol. II, Madrid
Ogden R, Linacre A (2015) Wildlife forensic science: a review of genetic geographic origin assignment. Forensic Sci Int Genet 18:152–159
Paetkau D, Calvert W, Stirling I, Strobeck C (1995) Microsatellite analysis of population structure in Canadian polar bears. Mol Ecol 4:347–354
Pandey D, Brown C (2000) Teak: a global overview. Unasylva 51(201):3–12
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Perera WRH (1962) The development of forest plantations in Ceylon since the seventeenth century. Ceylon Forester 5:142–147
Persson A (1971) Observations from a provenance trial of Tectona grandis Linn. f. at Longuza, Tanga region. Tanzania Silviculture Res Note 22:1–13
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) GENECLASS2: a software for genetic assignment and first-generation migrant detection. J Hered 95:536–539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2010) Documentation for structure software: version 2.3—February 2, 2010. Department of Human Genetics, University of Chicago
Queller DC, Goodnight KF (1989) Estimating relatedness using molecular markers. Evolution 43:258–275
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci U S A 94:9197–9201
Ritland K (1996) Estimators for pairwise relatedness and inbreeding coefficients. Genet Res 67:175–186
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Saitou N, Nei M (1987) The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Tariel J (1966) Le Teck en Côte-d’Ivoire. Bois For Trop 107:27–47
The International Plant Names Index (2014) Published on the Internet http://www.ipni.org. Accessed 31 Jan 2014
Thwe-Thwe-Win, Hirao T, Watanabe A, Goto S (2015) Current genetic structure of teak (Tectona qrandis) in Myanmar based on newly developed chloroplast single nucleotide polymorphism and nuclear single sequence repeat markers. Trop Conserv Sci 8:235–256
Thwe-Thwe-Win, Hirao T, Goto S (2016) Genetic composition of exotic and native teak (Tectona grandis) in Myanmar as revealed by cpSNP and nrSSR markers. Conserv Genet 17:251–258
Tnah LH, Lee SL, Ng KKS, Faridah QZ, Faridah-Hanum I (2010) Forensic DNA profiling of tropical timber species in Peninsular Malaysia. Forest Ecol Manag 259:1436–1446
Verhaegen D, Ofori D, Fofana I, Poitel M, Vaillant A (2005) Development and characterization of microsatellite markers in Tectona grandis (Linn. f). Mol Ecol Notes 5:945–947
Verhaegen D, Fofana IJ, Logossa ZA, Ofori D (2010) What is the genetic origin of teak (Tectona grandis L.) introduced in Africa and in Indonesia? Tree Genet Genomes 6:717–733
Wang J (2007) Triadic IBD coefficients and applications to estimating pairwise relatedness. Genet Res 89:135–153
Wang J (2011) COANCESTRY: a program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol Ecol Resour 11(1):141–145
Wood PJ (1967) Teak planting in Tanzania. Proceedings from FAO world symposium on man-made forests. Document 3:1631–1644. Canberra, Australia
Acknowledgments
We are grateful to Lars Schmidt for having conducted some of the field sampling in the provenance trial in Longuza, Tanzania. We would also like to thank Mr. Wiroj and his staff from the Royal Forest Department for excellent help during the collection of leaf samples in the field trial in Pha Nak Khao, Thailand. At the same time, our thanks go to Nutthakorn Semsantud and Verapong Suangtho for invaluable help with logistics and collection in Thailand. We further acknowledge the contribution of the different members of CAMCORE who provided us with samples of landraces from Latin America, Indonesia and Africa. The International Woodland Company (IWC) supplied us with samples from their two plantations in Panama. During the various steps of seed handling, germination and genotyping, we had good help of Kristin Marie Lassen, Kirstine Bräuner Nielsen and Jesper Vind. This research was supported by the Danish International Development Agency (Danida), and the final steps of analyses and publishing of the results were supported by the GB Hartmann Family Foundation. Finally, we acknowledge the comments from three anonymous reviewers which improved the final version of this manuscript.
Data archiving statement
The data—consisting of genotypic data for six SSRs in 721 individuals from 29 provenances as well as in 445 individuals from the 17 landraces—will be archived in Dryad (http://datadryad.org/).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Z. Kaya
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hansen, O.K., Changtragoon, S., Ponoy, B. et al. Worldwide translocation of teak—origin of landraces and present genetic base. Tree Genetics & Genomes 13, 87 (2017). https://doi.org/10.1007/s11295-017-1170-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-017-1170-8