Abstract
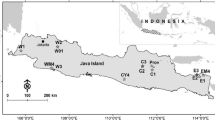
With the rapid fragmentation of tropical forests harboring valuable tree species, conservation of natural genetic resources is an important issue. In Myanmar, teak plantations have been established by Myanmar government since the 1700 s using local Myanmar teak. Commercial plantations have recently been established by the private sector using both exotic and Myanmar teak without consideration of their genetic make-up. If the genetic composition of commercial teak plantations is severely different from that of Myanmar teak, introgression of non-indigenous genes could damage the remaining natural populations. We investigated genetic compositions of commercial plantations using both exotic and Myanmar teak seeds with 10 nuclear simple sequence repeat and three chloroplast single nucleotide polymorphism markers. We then compared the genetic compositions of these populations with those of neighboring native teak forests. The genetic diversity and composition of one exotic plantation using Costa Rican seeds was similar to those of native populations. However, the diversity of the other three exotic plantations was low and their composition was markedly different from those of native populations. Our results suggest that exotic gene flow would cause serious genetic disturbance. Commercial plantations using Myanmar seeds were characterized by relatively high genetic diversity and by many genetic components. These results suggest that these plantations may be established using various seed sources in Myanmar. Given that native teak in Myanmar is geographically structured, native gene pools will be homogenized by gene flow from these commercial plantations. Seed transfer guidelines based on genetic information should be considered in future.



Similar content being viewed by others
References
Evanno GS, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Goudet J (2001) FSTAT version 2.9.3. A program to estimate and test gene diversities and fixation indices. http://www2.unil.ch/popgen/softwares/fstat.htm
Hufford KM, Mazer SJ (2003) Plant ecotypes: genetic differentiation in the age of ecological restoration. Trends Ecol Evol 18:147–155
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Laikre L, Schwartz MK, Waples RS, Ryman N, The GeM Working Group (2010) Compromising genetic diversity in the wild: unmonitored large-scale release of plants and animals. Trends Ecol Evol 25:520–529
McKay JK, Christian CE, Harrison CE (2005) How local is local? A review of practical and conceptual issue in the genetic of restoration. Restor Ecol 13:432–440
Montalvo AM, Williams SL, Rice SL, Buchmann SL, Cory C, Handel SN, Nabhan GP, Primack R, Robichaus RH (1997) Restoration biology: a population biology perspective. Restor Ecol 5:277–290
Minn Y, Prinz K, Finkeldey R (2014) Genetic variation of teak (Tectona grandis Linn. f.) in Myanmar revealed by microsatellites. Tree Genet Genomes 10:1435–1449
Morgenstern EK (1996) Geographic variation in forest trees. UBC Press, Vancouver. ISBN 0-7748-0579-X
Pandey D, Brown C (2000) Teak: a global overview. Unasylva 201:3–13
Prabha SS, Indira EP, Nair PN (2007) Contemporary gene flow in natural teak forest of Kerala using microsatellite markers. In: Proceedings 19th Kerala Science Congress 2007, pp 142–144
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Riina J, Thong HL, Leong LS, Judy L, Laura S (2014) Integrating genetic factors into management of tropical Asian production forests: a review of current knowledge. For Ecol Manag 315:191–201
Robledo-Arnuncio JJ, Navascués M, González-Martínez SC, Gil L (2009) Estimating gametic introgression rates in a risk assessment context: a case study with Scots pine relicts. Heredity 103:385–393
Rosenberg NA (2007) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Shiraishi S, Watanabe A (1995) Identification of chloroplast genome between Pinus densiflora Sieb. et Zucc. and P. thunbergii Parl. based on the polymorphism in rbcL gene. J Jpn For Soc 77:429–436 (in Japanese with English summary)
Steinitz O, Robledo-Arnuncio JJ, Nathan R (2012) Effects of forest plantations on the genetic composition of conspecific native Aleppo pine populations. Mol Ecol 21:300–313
Thwe-Thwe-Win, Watanabe A, Hirao T, Isoda K, Ishizuka W, Goto S (2015a) Genetic diversity of teak populations in native regions and plantations in Myanmar detected by microsatellite markers. Bull Uni Tokyo For 132:1–15
Thwe-Thwe-Win, Hirao T, Watanabe A, Goto S (2015b) Current genetic structure of teak (Tectona grandis) in Myanmar based on newly developed chloroplast single nucleotide polymorphism and nuclear simple sequence repeat markers. Trop Conserv Sci 8:235–256
Unger GM, Vendramin GG, Robledo-Arnuncio JJ (2014) Estimating exotic gene flow into native pine stands: zygotic vs. gametic components. Mol Ecol 23:5435–5447
Verhaegen D, Ofori D, Fofana IJ, Poitel M, Vaillant A (2005) Development and characterization of microsatellite markers in Tectona grandis (Linn.f). Mol Ecol 5:945–947
Acknowledgments
We thank Chunlan Lian for sharing experimental facilities. For helpful advice, we thank Wataru Ishizuka, Mineaki Aizawa, Kentarou Uchiyama, and Naoko Miura. We acknowledge U Zaw Win Myint, Director of the Forest Research Institute, Myanmar, U Aye Thiha, U Than Oo, Daw Khin Thida Tun, and Daw Soe Soe New for helping in the collection of exotic teak samples.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thwe-Thwe-Win, Hirao, T. & Goto, S. Genetic composition of exotic and native teak (Tectona grandis) in Myanmar as revealed by cpSNP and nrSSR markers. Conserv Genet 17, 251–258 (2016). https://doi.org/10.1007/s10592-015-0777-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-015-0777-2