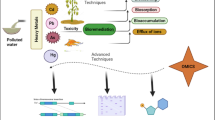
Abstract
The rapid industrial revolution significantly increased heavy metal pollution, becoming a major global environmental concern. This pollution is considered as one of the most harmful and toxic threats to all environmental components (air, soil, water, animals, and plants until reaching to human). Therefore, scientists try to find a promising and eco-friendly technique to solve this problem i.e., bacterial bioremediation. Various heavy metal resistance mechanisms were reported. Omics technologies can significantly improve our understanding of heavy metal resistant bacteria and their communities. They are a potent tool for investigating the adaptation processes of microbes in severe conditions. These omics methods provide unique benefits for investigating metabolic alterations, microbial diversity, and mechanisms of resistance of individual strains or communities to harsh conditions. Starting with genome sequencing which provides us with complete and comprehensive insight into the resistance mechanism of heavy metal resistant bacteria. Moreover, genome sequencing facilitates the opportunities to identify specific metal resistance genes, operons, and regulatory elements in the genomes of individual bacteria, understand the genetic mechanisms and variations responsible for heavy metal resistance within and between bacterial species in addition to the transcriptome, proteome that obtain the real expressed genes. Moreover, at the community level, metagenome, meta transcriptome and meta proteome participate in understanding the microbial interactive network potentially novel metabolic pathways, enzymes and gene species can all be found using these methods. This review presents the state of the art and anticipated developments in the use of omics technologies in the investigation of microbes used for heavy metal bioremediation.




Similar content being viewed by others
Data availability
No datasets were generated or analysed during the current study.
References
Abbaszade G, Szabó A, Vajna B, Farkas R, Szabó C, Tóth E (2020) Whole genome sequence analysis of Cupriavidus campinensis S14E4C, a heavy metal resistant bacterium. Mol Biol Rep 47:3973–3985. https://doi.org/10.1007/s11033-020-05490-8
Abdelhaleem HA, Zein HS, Azeiz A, Sharaf AN, Abdelhadi AA (2019) Identification and characterization of novel bacterial polyaromatic hydrocarbon-degrading enzymes as potential tools for cleaning up hydrocarbon pollutants from different environmental sources. Environ Toxicol Pharmacol 7(2):108–116. https://doi.org/10.1016/j.etap.2019.02.009
Acharya C, Khare D, Divya TV, Chandwadkar P, Kolhe N (2021) Role of biological macromolecules of microbes in metal bioremediation. In: Microbial and Natural Macromolecules. United States: Academic Press 497–529.https://doi.org/10.1016/b978-0-12-820084-1.00020-x
Afridi MS, Van Hamme JD, Bundschuh J, Sumaira Khan MN, Salam A, Chaudhary HJ (2021) Biotechnological approaches in agriculture and environmental management-bacterium Kocuria rhizophila 14ASP as heavy metal and salt-tolerant plant growth-promoting strain. Biologia 76(10):3091–3105. https://doi.org/10.1007/s11756-021-00826-6
Ahad RIA, Syiem MB (2018) Copper-induced morphological, physiological and biochemical responses in the cyanobacterium Nostoc muscorum Meg 1. Nat Environ Pollut Technol 17(4):1077–1086. https://doi.org/10.1007/s13205-017-0730-9
Aminov RI (2011) Horizontal gene exchange in environmental microbiota. Front Microbiol 2:158. https://doi.org/10.3389/fmicb.2011.00158
Arsène-Ploetze F, Koechler S, Marchal M, Coppée JY, Chandler M, Bonnefoy V, Bertin PN (2010) Structure, function, and evolution of the Thiomonas spp. genome. PLoS Genet 6(2):e1000859. https://doi.org/10.1371/journal.pgen.1000859
Ayangbenro AS, Babalola OO (2017) A new strategy for heavy metal polluted environments: a review of microbial biosorbents. Int J Environ Res 14(1):94–109. https://doi.org/10.3390/ijerph14010094
Aziz RK, Bartels D, Best A, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Zagnitko O (2008) The RAST server: Rapid annotations using subsystems technology. BMC Genom 9:1–15. https://doi.org/10.1186/1471-2164-9-75
Baggerly KA, Morris JS, Edmonson SR, Coombes KR (2005) Signal in noise: evaluating reported reproducibility of serum proteomics tests for ovarian cancer. J Natl Cancer Inst 97:307–309. https://doi.org/10.1093/jnci/dji008
Baksh KA, Zamble DB (2020) Allosteric control of metal-responsive transcriptional regulators in bacteria. J Biol Chem 295(6):1673–1684. https://doi.org/10.1074/jbc.rev119.011444
Balali-Mood M, Naseri K, Tahergorabi Z, Khazdair MR, Sadeghi M (2021) Toxic mechanisms of five heavy metals: mercury, lead, chromium, cadmium, and arsenic. Front Pharmacol 2(2):227–240. https://doi.org/10.1016/j.enmm.2020.100283
Ballester LY, Luthra R, Kanagal-Shamanna R, Singh RR (2016) Advances in clinical next-generation sequencing: target enrichment and sequencing technologies. Expert Rev Mol Diagn 16:357–372. https://doi.org/10.1586/14737159.2016.1133298
Battistuzzi FU, Hedges SB (2009) A major clade of prokaryotes with ancient adaptations to life on land. Mol Biol Evol 26(2):335–343. https://doi.org/10.1093/molbev/msn247
Baya G, Muhindi S, Ngendahimana V, Caguiat J (2021) Potential whole-cell biosensors for detection of metal using merr family proteins from Enterobacter sp. Ysu and Stenotrophomonas maltophilia or02. Micromachines 12(2): 142. https://doi.org/10.3390/mi12020142
Beck BR, Park GS, Lee YH, Im S, Jeong DY, Kang J (2019) Whole genome analysis of Lactobacillus plantarum strains isolated from Kimchi and determination of probiotic properties to treat mucosal infections by Candida albicans and Gardnerella vaginalis. Front Microbiol 10:433. https://doi.org/10.3389/fmicb.2019.00433
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2008) GenBank. Nucleic Acids Res 36:D25–D30. https://doi.org/10.1093/nar/gkm929
Bertin PN, Heinrich-Salmeron A, Pelletier E, Goulhen-Chollet F, Arsène-Ploetze F, Gallien S, Le Paslier D (2011) Metabolic diversity among main microorganisms inside an arsenic-rich ecosystem revealed by meta-and proteo-genomics. ISME J 5(11):1735–1747. https://doi.org/10.1038/ismej.2011.51
Besser J, Carleton HA, Gerner-Smidt P, Lindsey RL, Trees E (2018) Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clin Microbiol Infect 24(4):335–341. https://doi.org/10.1016/j.cmi.2017.10.013
Bhaskar PV, Bhosle NB (2006) Bacterial extracellular polymeric substance (EPS): a carrier of heavy metals in the marine food-chain. Environ Int 32:191–198. https://doi.org/10.1016/j.envint.2005.08.010
Bordel S, Martín-González D, Muñoz R, Santos-Beneit F (2023) Genome sequence analysis and characterization of Bacillus altitudinis B12, a polylactic acid-and keratin-degrading bacterium. Mol Genet Genomics 298(2):389–398. https://doi.org/10.1007/s00438-022-01989-w
Branca JJV, Morucci G, Pacini (2018) A cadmium-induced neurotoxicity: still much ado. Neural Regen Res 13(11):1879. https://doi.org/10.4103/1673-5374.239434
Briffa J, Sinagra E, Blundell R (2020) Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon 6(9):e04691. https://doi.org/10.1016/j.heliyon.2020.e04691
Brown NL, Stoyanov JV, Kidd SP, Hobman JL (2003) The MerR family of transcriptional regulators. FEMS Microbiol Rev 27(3):145–163. https://doi.org/10.1016/S0168-6445(03)00051-2
Bruins MR, Kapil S, Oehme FW (2000) Microbial resistance to metals in the environment. Ecotoxicol Environ Saf 45(3):198–207. https://doi.org/10.1006/eesa.1999.1860
Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick T, Inns, Floto RA (2013) Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381(9877):1551–1560. https://doi.org/10.1016/s0140-6736
Busenlehner LS, Cosper NJ, Scott RA, Rosen BP, Wong MD, Giedroc DP (2001) Spectroscopic properties of the metalloregulatory cd (II) and pb (II) sites of S. Aureus pI258 CadC. Biochemistry 40(14):4426–4436. https://doi.org/10.1021/bi010006g
Bustamante P, Covarrubias PC, Levicán G, Katz A, Tapia P, Holmes D, Quatrini R, Orellana O (2012) ICE Afe 1, an actively excising genetic element from the biomining bacterium Acidithiobacillus ferrooxidans. J Mol Microbiol Biotechnol 22:399–407. https://doi.org/10.1159/000346669
Cao K, Li S, Wang Y, Hu H, Xiang S, Zhang Q, Liu Y (2023) Cellular uptake of nickel by NikR is regulated by phase separation. Cell Rep 42(6):112518. https://doi.org/10.1016/j.celrep.2023.112518
Capdevila DA, Edmonds KA, Giedroc DP (2017) Metallochaperones and metalloregulation in bacteria. Essays Biochem 61(2):177–200. https://doi.org/10.1042/ebc20160076
Cárdenas JP, Valdés J, Quatrini R, Duarte F, Holmes DS (2010) Lessons from the genomes of extremely acidophilic bacteria and archaea with special emphasis on bioleaching microorganisms. Appl Microbiol Biotechnol 88:605–620. https://doi.org/10.1007/s00253-010-2795-9
Carro L, Veyisoglu A, Guven K, Schumann P, Klenk HP, Sahin N (2022) Genomic analysis of a novel heavy metal resistant isolate from a Black Sea contaminated sediment with the potential to degrade alkanes: Plantactinospora alkalitolerans sp. nov. Diversity 14:947. https://doi.org/10.3390/d14110947
Chen P, Greenberg B, Taghavi S, Romano C, van der Lelie D, He C (2005) An exceptionally selective lead (II) regulatory protein from Ralstonia metallidurans: development of a fluorescent lead (II) probe. Angew Chem 117(18):2775–2779. https://doi.org/10.1002/ange.200462443
Chen LX, Hu M, Huang LN, Hua ZS, Kuang JL, Li SJ, Shu WS (2015) Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage. ISME J 9(7):1579–1592. https://doi.org/10.1038/ismej.2014.245
Chen Y, Jiang Y, Huang H, Mou L, Ru J, Zhao J, Xiao S (2018) Long-term and high-concentration heavy-metal contamination strongly influences the microbiome and functional genes in Yellow River sediments. Sci Total Environ 637:1400–1412. https://doi.org/10.1016/j.scitotenv.2018.05.109
Chen L, Zhang X, Zhang M, Zhu Y, Zhuo R (2022) Removal of heavy-metal pollutants by white rot fungi: mechanisms, achievements, and perspectives. J Clean Prod 354:131681. https://doi.org/10.1016/j.jclepro.2022.131681
Chen Y, Liu X, Li Q, Cai X, Wu W, Wu Q, Yuan W, Deng X, Liu Z, Zhao S, Wang B (2023) Integrated genomics and transcriptomics reveal the extreme heavy metal tolerance and adsorption potentiality of Staphylococcus equorum. Int J Biol Macromol 229:388–400. https://doi.org/10.1016/j.ijbiomac.2022.12.298
Childers SE, Ciufo S, Lovley DR (2002) Geobacterm etallireducens accesses insoluble Fe (III) oxide by chemotaxis. Nature 416(6882): 767-9. https://doi.org/10.1038/416767a
Chivers PT, Sauer RT (2000) Regulation of high affinity nickel uptake in bacteria: Ni2+–dependent interaction of NikR with wild-type and mutant operator sites. J Biol Chem 275(26):19735–19741. https://doi.org/10.1074/jbc.m002232200
Choudhury R, Srivastava S (2001) Zinc resistance mechanisms in bacteria. Curr Microbiol 43:316–321. https://doi.org/10.1007/s002840010309
Clemens S, Simm C (2003) Schizosaccharomy Cespombe as a model for metal homeostasis in plant cells: the phytochelatin-dependent pathway is the main cadmium detoxification mechanism. New Phytol 159(2):323–330. https://doi.org/10.1046/j.1469-8137.2003.00811.x
Coombs JM, Barkay T (2005) New findings on evolution of metal homeostasis genes: evidence from comparative genome analysis of bacteria and archaea. Appl Environ Microbiol 71(11):7083–7091. https://doi.org/10.1128/aem.71.11.7083-7091.2005
Dash HR, Basu S, Das S (2017) Evidence of mercury trapping in biofilm-EPS and mer operon-based volatilization of inorganic mercury in a marine bacterium Bacillus cereus BW-201B. Arch Microbiol 199:445–455. https://doi.org/10.1007/s00203-016-1317-2
Denef VJ, Kalnejais LH, Mueller RS, Wilmes P, Baker BJ, Thomas BC, Banfield JF (2010) Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proc Natl Acad Sci 107(6):2383–2390. https://doi.org/10.1073/pnas.0907041107
Deng S, Zhang X, Zhu Y, Zhuo R (2024) Recent advances in phyto-combined remediation of heavy metal pollution in soil. Biotechnol Adv 108337. https://doi.org/10.1016/j.biotechadv.2024.108337
Deochand DK, Grove A (2017) MarR family transcription factors: dynamic variations on a common scaffold. Crit Rev Biochem Mol Biol 52(6):595–613. https://doi.org/10.1080/10409238.2017.1344612
Dobrindt U, Hochhut B, Hentschel U, Hacker J (2004) Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2(5):414–424. https://doi.org/10.4067/s0716-97602013000400008
Dopson M (2016) Physiological and phylogenetic diversity of acidophilic bacteria. Acidophiles: life in extremely acidic environments. Academic, United Kingdom, pp 79–92. https:/ doi: https://doi.org/10.21775/9781910190333.05
Dzidic S, Bedeković V (2003) Horizontal gene transfer-emerging multidrug resistance in hospital bacteria. Acta Pharmacol Sin 24(6):519–526
El Zowalaty ME, Falgenhauer L, Forsythe S, Helmy YA (2023) Draft genome sequences of rare Lelliottia nimipressuralis strain MEZLN61 and two Enterobacter kobei strains MEZEK193 and MEZEK194 carrying mobile colistin resistance gene mcr-9 isolated from wastewater in South. J Glob Antimicrob Resist 33:231–237. https://doi.org/10.1016/j.jgar.2023.03.007
El-Beltagi HS, Halema AA, Almutairi ZM, Almutairi HH, Elarabi NI, Abdelhadi AA, Abdelhaleem HA (2024) Draft genome analysis for Enterobacter kobei, a promising lead bioremediation bacterium. Front Bioeng Biotechnol 11:1335854. https://doi.org/10.3389/fbioe.2023.1335854
El-Helow ER, Sabry SA, Amer RM (2000) Cadmium biosorption by a cadmium resistant strain of Bacillus thuringiensis: regulation and optimization of cell surface affinity for metal cations. Biometals 13:273–280. https://doi.org/10.1023/a:1009291931258
Elarabi NI, Abdelhadi AA, Ahmed RH, Saleh I, Arif IA, Osman G, Ahmed DS (2020) Bacillus aryabhattai FACU: a promising bacterial strain capable of manipulate the glyphosate herbicide residues. Saudi J Biol Sci 27(9):2207–2214. https://doi.org/10.1016/j.sjbs.2020.06.050
Elarabi NI, Halema AA, Abdelhadi AA, Henawy AR, Samir O, Abdelhaleem HA (2023) Draft genome of Raoultella planticola, a high lead resistance bacterium from industrial wastewater. AMB Express 13(1):14–30. https://doi.org/10.1186/s13568-023-01519-w
Engin AB, Engin ED, Engin A (2023) Effects of co-selection of antibiotic-resistance and metal-resistance genes on antibiotic-resistance potency of environmental bacteria and related ecological risk factors. Environ Toxicol Pharmacol 98:104081. https://doi.org/10.1016/j.etap.2023.104081
Fan X, Tang J, Nie L, Huang J, Wang G (2018) High-quality-draft genome sequence of the heavy metal resistant and exopolysaccharides producing bacterium Mucilaginibacter Pedocola TBZ30 T. Stand Genom Sci 13:1–8. https://doi.org/10.1186/s40793-018-0337-8
Fang J, Jiao X, Cheng H, Tang B, Wang W, Yu T, Jiang H (2021) Draft genome of a multidrug and multi-heavy metal resistant Vibrio parahaemolyticus ST165 strain of Penaeus vannamei from seawater farms in Zhejiang, China. J Glob Antimicrob Resist 26:323–325. https://doi.org/10.1016/j.jgar.2021.07.015
Fariq A, Blazier JC, Yasmin A, Gentry TJ, Deng Y (2019) Whole genome sequence analysis reveals high genetic variation of newly isolated Acidithiobacillus ferrooxidans IO-2 C. Sci Rep 9(1):13049. https://doi.org/10.1038/s41598-019-49213-x
Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, Klimke W (2019) Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 63(11):10–1128. https://doi.org/10.1128/aac.00483-19
Gao X, Wei M, Zhang X, Xun Y, Duan M, Yang Z, Zhu M, Zhu Y, Zhuo R (2024) Copper removal from aqueous solutions by white rot fungus Pleurotus Ostreatus GEMB-PO1 and its potential in co-remediation of copper and organic pollutants. Bioresour Technol 395:130337. https://doi.org/10.1016/j.biortech.2024.130337
Gautam PK, Gautam RK, Banerjee S, Chattopadhyaya MC, Pandey JD (2016) Heavy metals in the environment: fate, transport, toxicity and remediation technologies. Heavy metals: sources, toxicity and remediation techniques. Nova Sci Publishers 101 – 30, United States. https://doi.org/10.1039/9781782620174-00001
Gibson MK, Forsberg KJ, Dantas G (2015) Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J 9(1):207–216
Große C, Anton A, Hoffmann T, Franke S, Schleuder G, Nies DH (2004) Identification of a regulatory pathway that controls the heavy-metal resistance system Czc via promoter czcNp in Ralstonia metallidurans. Arch Microbiol 182:109–118. https://doi.org/10.1007/s00203-004-0670-8
Guo T, Lou C, Zhai W, Tang X, Hashmi MZ, Murtaza R, Xu J (2018) Increased occurrence of heavy metals, antibiotics and resistance genes in surface soil after long-term application of manure. Sci Total Environ 635:995–1003. https://doi.org/10.1016/j.scitotenv.2018.04.194
Hallberg KB, Hedrich S, Johnson DB (2011) Acidiferrobacter thiooxydans, gen nov sp nov, an acidophilic, thermo-tolerant, facultatively anaerobic iron-and sulfur-oxidizer of the family Ectothiorhodospiraceae. Extremophiles 15:271–279. https://doi.org/10.1007/s00792-011-0359-2
Hao X, Zhu J, Rensing C, Liu Y, Gao S, Chen W, Liu YR (2021) Recent advances in exploring the heavy metal (loid) resistant microbiome. Comput Struct Biotechnol J 19:94–109. https://doi.org/10.1016/j.csbj.2020.12.006
Hasanein P, Emamjomeh A (2019) Beneficial effects of natural compounds on heavy metal–induced hepatotoxicity. In Dietary interventions in liver disease, United States: Academic Press 345 – 55. https://doi.org/10.1016/b978-0-12-814466-4.00028-8
Heuer H, Smalla K (2012) Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol Rev 36(6):1083–1104. https://doi.org/10.1111/j.1574-6976.2012.00337.x
Hong KW, Thinagaran DA, Gan HM, Yin WF, Chan KG (2012) Whole-genome sequence of Cupriavidus sp. strain BIS7, a heavy-metal-resistant bacterium. Am Soc Microbiol 194(22):6324–6324. https://doi.org/10.1128/jb.01608-12
Huang HT, Maroney MJ (2019) Ni (II) sensing by RcnR does not require an FrmR-like intersubunit linkage. Inorg Chem 58(20):13639–13653. https://doi.org/10.1021/acs.inorgchem.9b01096
Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13. https://doi.org/10.1093/nar/gkn923
Huo YY, Li ZY, Cheng H, Wang CS, Xu XW (2014) High quality draft genome sequence of the heavy metal resistant bacterium Halomonas zincidurans type strain B6 T. Stand Genom Sci 9(2):1–9. https://doi.org/10.1186/1944-3277-9-30
Igiri BE, Okoduwa SI, Idoko GO, Akabuogu EP, Adeyi AO, Ejiogu IK (2018) Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: a review. J Toxicol 2018:1–16. https://doi.org/10.1155/2018/2568038
Irawati W, Parhusip AJ, Sopiah N (2015) Heavy metals biosorption by copper resistant bacteria of Acinetobacter Sp. IrC2. Microbiol Indonesia 9:163–170. https://doi.org/10.5454/mi.9.4.4
Istiaq A, Shuvo MSR, Rahman KMJ, Siddique MA, Hossain MA, Sultana M (2019) Adaptation of metal and antibiotic resistant traits in novel β-Proteobacterium Achromobacter xylosoxidans BHW-15. Peer J 7:e6537. https://doi.org/10.7717/peerj.6537
Iyer A, Mody K, Jha B (2004) Accumulation of hexavalent chromium by an exopolysaccharide producing marine enterobactercloaceae. Mar Pollut Bull 49(11–12):974–977. https://doi.org/10.1016/j.marpolbul.2004.06.023
Izrael-Živković L, Beškoski V, Rikalović M, Kazazić S, Shapiro N, Woyke T, Karadžić I (2019) High-quality draft genome sequence of Pseudomonas aeruginosa san ai, an environmental isolate resistant to heavy metals. Extremophiles 23:399–405. https://doi.org/10.1007/s00792-019-01092-w
Jan R, Khan MA, Asaf S, Lee IJ, Kim KM (2019) Metal resistant endophytic bacteria reduces cadmium, nickel toxicity, and enhances expression of metal stress related genes with improved growth of Oryza sativa, via regulating its antioxidant machinery and endogenous hormones. Plants 8(10):363–380. https://doi.org/10.3390/plants8100363
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. https://doi.org/10.1093/nar/gkn201
Johnson H, Cho H, Choudhary M (2019) Bacterial heavy metal resistance genes and bioremediation potential. Comput Mol Biol 9(1):1–12. https://doi.org/10.4236/cmb.2019.91001
Joshi S, Gangola S, Bhandari G, Bhandari NS, Nainwal D, Rani A, Malik S, Slama P (2023) Rhizospheric bacteria: the key to sustainable heavy metal detoxification strategies. Front Microbiol 14:1229828. https://doi.org/10.3389/fmicb.2023.1229828
Jung J, Lee SJ (2019) Biochemical and biodiversity insights into heavy metal ion-responsive transcription regulators for synthetic biological heavy metal sensors. J Microbiol Biotechnol 29(10):1522–1542. https://doi.org/10.4014/jmb.1908.08002
Kandari D, Joshi H, Bhatnagar R (2021) Zur: zinc-sensing transcriptional regulator in a diverse set of bacterial species. Pathogens 10(3):344. https://doi.org/10.3390/pathogens10030344
Kang SM, Asaf S, Khan AL, Lubna, Khan A, Mun BG, Khan MA, Gul H, Lee IJ (2020) Complete genome sequence of Pseudomonas psychrotolerans CS51, a plant growth-promoting bacterium, under heavy metal stress conditions. Microorganisms 8:382. https://doi.org/10.3390/microorganisms8030382
Khaldi N, Collemare J, Lebrun MH, Wolfe KH (2008) Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol 9:1–10. https://doi.org/10.1186/gb-2008-9-1-r18
Khaleque HN, Corbett MK, Ramsay JP, Kaksonen AH, Boxall NJ, Watkin EL (2017) Complete genome sequence of Acidihalobacter prosperus strain F5, an extremely acidophilic, iron-and sulfur-oxidizing halophile with potential industrial applicability in saline water bioleaching of chalcopyrite. J Biotech 262:56–59. https://doi.org/10.1016/j.jbiotec.2017.10.001
Klonowska A, Moulin L, Ardley JK, Braun F, Gollagher MM, Zandberg JD, Reeve WG (2020) Novel heavy metal resistance gene clusters are present in the genome of Cupriavidus Neocaledonicus STM 6070, a new species of Mimosa pudica microsymbiont isolated from heavy-metal-rich mining site soil. BMC Genom 21(1):1–18. https://doi.org/10.1186/s12864-020-6623-z
Kumar A, Bisht BS, Joshi VD, Dhewa T (2011) Review on bioremediation of polluted environment: a management tool. Int J Environ Sci 1(6):1079–1093
Kumari S, Jamwal R, Mishra N, Singh DK (2020) Recent developments in environmental mercury bioremediation and its toxicity: Areview. Environ Nanotechnol Monit Manag 13(1):100–133. https://doi.org/10.1016/j.enmm.2020.100283
Lancaster SM, Sanghi A, Wu S, Snyder MP (2020) A customizable analysis flow in integrative multi-omics. Biomolecules 10(12):1606. https://doi.org/10.3390/biom10121606
Land M, Hauser L, Jun SR, Nookaew I, Leuze MR, Ahn TH, Ussery DW (2015) Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics 15:141–161. https://doi.org/10.1007/s10142-015-0433-4
Lee SW, Glickmann E, Cooksey DA (2001) Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator. Appl Environ Microbiol 67(4):1437–1444. https://doi.org/10.1128/aem.67.4.1437-1444.2001
Lee H, Gurtowski J, Yoo S, Nattestad M, Marcus S, Goodwin S, Schatz MC (2016) Third-generation sequencing and the future of genomics. BioRxiv 048603. https://doi.org/10.1101/048603
Leyn SA, Rodionov DA (2015) Comparative genomics of DtxR family regulons for metal homeostasis in Archaea. J Bacterial 197(3):451–458. https://doi.org/10.1128/jb.02386-14
Li M, Wen J (2021) Recent progress in the application of omics technologies in the study of bio-mining microorganisms from extreme environments. Microb Cell Factories 20(1):1–11. https://doi.org/10.1186/s12934-021-01671-7
Li LG, Xia Y, Zhang T (2017) Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J 11(3):651–662. https://doi.org/10.1038/ismej.2016.155
Li L, Liu Z, Meng D, Liu X, Li X, Zhang M, Yin H (2019) Comparative genomic analysis reveals the distribution, organization, and evolution of metal resistance genes in the genus Acidithiobacillus. Appl Environ Microbiol 85(2):e02153–e02118. https://doi.org/10.1128/aem.02153-18
Li L, Wang S, Shen X, Jiang M (2020a) Ecological risk assessment of heavy metal pollution in the water of China’s coastal shellfish culture areas. Environ Sci Pollut Res 27:8392–8402. https://doi.org/10.1007/s11356-020-08173-w
Li S, Zhao B, Jin M, Hu L, Zhong H, He Z (2020b) A comprehensive survey on the horizontal and vertical distribution of heavy metals and microorganisms in soils of a Pb/Zn smelter. J Hazard Mater 400:123255. https://doi.org/10.1016/j.jhazmat.2020.123255
Li IC, Yu GY, Huang JF, Chen ZW, Chou CH (2022a) Comparison of Reference-Based Assembly and De Novo Assembly for bacterial plasmid Reconstruction and AMR Gene Localization in Salmonella enterica Serovar Schwarzengrund isolates. Microorganisms 10(2):227. https://doi.org/10.3390/microorganisms10020227
Li Z, Henawy AR, Halema AA, Fan Q, Duanmu D, Huang RA (2022b) Wild Rice Rhizobacterium Burkholderia cepacia BRDJ enhances Nitrogen Use Efficiency in Rice. Int J Mol Sci 23:10769. https://doi.org/10.3390/ijms231810769
Li XL, Lv XY, Ji JB, Wang WD, Wang J, Wang C, Liu TL (2023) Complete genome sequence of Nguyenibacter sp. L1, a phosphate solubilizing bacterium isolated from Lespedeza bicolor rhizosphere. Front Microbiol 4:1257442. https://doi.org/10.3389/fmicb.2023.1257442
Lima AIG, Corticeiro SC, de Almeida EM, Figueira P (2006) Glutathione-mediated cadmium sequestration in Rhizobium leguminosarum. Enzyme Microb Technol 39:763–769. https://doi.org/10.1016/j.enzmictec.2005.12.009
Liu L, Li Y, Li S, Hu N, He Y, Pong R, Law M (2012) Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012:251364. https://doi.org/10.1201/b16568-3
Loman NJ, Pallen MJ (2015) Twenty years of bacterial genome sequencing. Nat Rev Microbiol 13(12):787–794. https://doi.org/10.1038/nrmicro3565
Lovely DR (2005) Improving functional analysis of genes relevant to environmental restoration via an analysis of the genome of Geobacter sulfurreducens. United States: UMASS Microbiol 2–5. https://doi.org/10.2172/895726
Lu H, Giordano F, Ning Z (2016) Oxford Nanopore MinION sequencing and genome assembly. Genom Proteom Bioinform 14(5):265–279. https://doi.org/10.1016/j.gpb.2016.05.004
Luo Y, Chen L, Lu Z, Zhang W, Liu W, Chen Y, Wang X, Du W, Luo J, Wu H (2022) Genome sequencing of biocontrol strain Bacillus amyloliquefaciens Bam1 and further analysis of its heavy metal resistance mechanism. Bioresour Bioprocess 9(1):74. https://doi.org/10.1186/s40643-022-00563-x
Luziatelli F, Ficca AG, Cardarelli M, Melini F, Cavalieri A, Ruzzi M (2020) Genome sequencing of Pantoea agglomerans C1 provides insights into molecular and genetic mechanisms of plant growth-promotion and tolerance to heavy metals. Microorganisms 8:153. https://doi.org/10.3390/microorganisms8020153
Mardis ER (2013) Next-generation sequencing platforms. Annu Rev Anal Chem 6:287–303. https://doi.org/10.1146/annurev-anchem-062012-092628
Margesin R, Plaza G, Kasenbacher S (2011) Characterization of bacterial communities at heavy-metal-contaminated sites. Chemosphere 82:1583–1588. https://doi.org/10.1016/j.chemosphere.2010.11.056
McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Wright GD (2013) The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57(7):3348–3357. https://doi.org/10.1128/aac.00419-13
Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, Van Gijsegem F (1985) Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol 62(1):328–334. https://doi.org/10.1128/jb.162.1.328-334.1985
Mohamed ZA (2001) Removal of cadmium and manganese by a non-toxic strain of the freshwater cyanobacterium Gloeothecemagna. Water Res 35(18):4405–4409. https://doi.org/10.1016/s0043-1354(01)00160-9
Mohamed MS, El-Arabi NI, El-Hussein A, El-Maaty SA, Abdelhadi AA (2020) Reduction of chromium-VI by chromium-resistant Escherichia coli FACU: a prospective bacterium for bioremediation. Folia Microbiol 65:687–696. https://doi.org/10.1007/s12223-020-00771-y
Mohan AK, Martis S, Chiplunkar S, Kamath S, Goveas LC, Rao CV (2019) Heavy metal tolerance of Klebsiella pneumoniae Kpn555 isolated from coffee pulp waste. Borneo J Resour Sci Technol 9:101–106. https://doi.org/10.33736/bjrst.v9i2
Mukhopadhyay R, Rosen BP, Phung LT, Silver S (2002) Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol Rev 26(3):311–325. https://doi.org/10.1111/j.1574-6976.2002.tb00617.x
Muñoz-García A, Arbeli Z, Boyacá-Vásquez V, Vanegas J (2022) Metagenomic and genomic characterization of heavy metal tolerance and resistance genes in the rhizosphere microbiome of Avicennia germinans in a semi-arid mangrove forest in the tropics. Mar Pollut Bull 184:114204. https://doi.org/10.1016/j.marpolbul.2022.114204
Nanda M, Kumar V, Sharma DK (2019) Multimetal tolerance mechanisms in bacteria: the resistance strategies acquired by bacteria that can be exploited to ‘clean-up’heavy metal contaminants from water. Aquat Toxicol 212:1–10. https://doi.org/10.1016/j.aquatox.2019.04.011
Narita M, Chiba K, Nishizawa H, Ishii H, Huang CC, Kawabata ZI, Endo G (2003) Diversity of mercury resistance determinants among Bacillus strains isolated from sediment of Minamata Bay. FEMS Microbiol Lett 223(1):73–82. https://doi.org/10.1016/s0378-1097(03)00325-2
Navarro CA, von Bernath D, Jerez CA (2013) Heavy metal resistance strategies of acidophilic bacteria and their acquisition: importance for biomining and bioremediation. Biol Res 46(4):363–371. https://doi.org/10.4067/s0716-97602013000400008
Nawaz MZ, Xu C, Qaria MA, Haider SZ, Khalid HR, Alghamdi HA, Zhu D (2023) Genomic and biotechnological potential of a novel oil-degrading strain Enterobacter kobei DH7 isolated from petroleum-contaminated soil. Chemosphere 340:139815. https://doi.org/10.1016/j.chemosphere.2023.139815
NCBI (2023) National Center for Biotechnology Information Genome Browser. http://www.ncbi.nlm.nih.gov/genome/browse [Accessed December 2023]
Nies DH, Rehbein G, Hoffmann T, Baumann C, Grosse C (2006) Paralogs of genes encoding metal resistance proteins in Cupriavidus metallidurans strain CH34. Microb Physiol 11(1–2):82–93. https://doi.org/10.1159/000092820
Ning M, Lo EH (2010) Opportunities and challenges in omics. Transl Stroke Res 1(4):233–237. https://doi.org/10.1007/s12975-010-0048-y
Orell A, Navarro CA, Arancibia R, Mobarec JC, Jerez CA (2010) Life in blue: copper resistance mechanisms of bacteria and archaea used in industrial biomining of minerals. Biotechnol Adv 28(6):839–848. https://doi.org/10.1016/j.biotechadv.2010.07.003
Orellana LH, Jerez CA (2011) A genomic island provides Acidithiobacillus ferrooxidans ATCC 53993 additional copper resistance: a possible competitive advantage. Appl Microbiol Biotechnol 92:761–767. https://doi.org/10.1007/s00253-011-3494-x
Pal C, Bengtsson-Palme J, Rensing C, Kristiansson E, Larsson DJ (2014) BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res 42(D1):D737–D743. https://doi.org/10.1093/nar/gkt1252
Pal A, Bhattacharjee S, Saha J, Sarkar M, Mandal P (2022) Bacterial survival strategies and responses under heavy metal stress: a comprehensive overview. Crit Rev Microbiol 48(3):327–355. https://doi.org/10.1080/1040841x.2021.1970512
Pardo R, Herguedas M, Barrado E, Vega M (2003) Biosorption of cadmium, copper, lead and zinc by inactive biomass of Pseudomonas putida. Anal Bioanal Chem 376:26–32. https://doi.org/10.1007/s00216-003-1843-z
Pattanapipitpaisal P, Brown NL, Macaskie LE (2001) Chromate reduction and 16S rRNA identification of bacteria isolated from a cr (VI)-contaminated site. Appl Microbiol Biotechnol 57(1–2) 257 – 61. https://doi.org/10.1007/s002530100758
Peng T, Ma L, Feng X, Tao J, Nan M, Liu Y, Zeng W (2017) Genomic and transcriptomic analyses reveal adaptation mechanisms of an Acidithiobacillus ferrivorans strain YL15 to alpine acid mine drainage. PLoS ONE 12(5):e0178008. https://doi.org/10.1371/journal.pone.0178008
Pervez MT, Abbas SH, Moustafa MF, Aslam N, Shah SSM (2022) A comprehensive review of performance of next-generation sequencing platforms. Biomed Res Int 2022:1–12. https://doi.org/10.1155/2022/3457806
Pinto E, Sigaud-kutner TC, Leitao MA, Okamoto OK, Morse D, Colepicolo P (2003) Heavy metal–induced oxidative stress in algae 1. J Phycol 39(6):1008–1018. https://doi.org/10.1111/j.0022-3646.2003.02-193.x
Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Gu Y (2013) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom 13(1):1–13. https://doi.org/10.1186/1471-2164-13-341
Rajendran S, Priya TAK, Khoo KS, Hoang TK, Ng HS, Munawaroh HSH, Show PL (2022) A critical review on various remediation approaches for heavy metal contaminants removal from contaminated soils. Chemosphere 287:132369. https://doi.org/10.1016/j.chemosphere.2021.132369
Ransohoff DF (2005) Lessons from controversy: ovarian cancer screening and serum proteomics. J Natl Cancer Inst 97:315–319. https://doi.org/10.1093/jnci/dji054
Rattan S, Zhou C, Chiang C, Mahalingam S, Brehm E, Flaws JA (2017) Exposure to endocrine disruptors during adulthood: consequences for female fertility. J Endocrinol 233(3):R109. https://doi.org/10.1530/joe-17-0023
Rehan M, Alhusays A, Serag AM, Boubakri H, Pujic P, Normand P (2022) The cadCA and cadB/DX operons are possibly induced in cadmium resistance mechanism by Frankia alni ACN14a. Electron J Biotechnol 60:86–96. https://doi.org/10.1016/j.ejbt.2022.09.006
Rensing C, Rosen BP (2000) Transport of soft metals in procaryotes. Molecular Biology and Toxicology of metals. CRC, United States, pp 129–150
Rensing C, Ghosh M, Rosen BP (1999) Families of soft-metal-ion-transporting ATPases. J Bacteriol 181(19):5891. https://doi.org/10.1128/jb.181.19.5891-5897.1999
Ridge PG, Zhang Y, Gladyshev VN (2008) Comparative genomic analyses of copper transporters and cuproproteomes reveal evolutionary dynamics of copper utilization and its link to oxygen. PLoS ONE 3(1):e1378–e1395. https://doi.org/10.1371/journal.pone.0001378
Saha RP, Samanta S, Patra S, Sarkar D, Saha A, Singh MK (2017) Metal homeostasis in bacteria: the role of ArsR–SmtB family of transcriptional repressors in combating varying metal concentrations in the environment. Biometals 30(4):459–503. https://doi.org/10.1007/s10534-017-0020-3
Saha J, Dey S, Pal A (2022) Whole genome sequencing and comparative genomic analyses of Pseudomonas aeruginosa strain isolated from arable soil reveal novel insights into heavy metal resistance and codon biology. Curr Genet 68(3–4):481–503. https://doi.org/10.1007/s00294-022-01245-z
Saini S, Dhania G (2020) Cadmium as an environmental pollutant: ecotoxicological effects, health hazards, and bioremediation approaches for its detoxification from contaminated sites. Bioremediation of industrial waste for environmental safety. Springer, Singapore. https://doi.org/10.1007/978-981-13-3426-9_15
Satam H, Joshi K, Mangrolia U, Waghoo S, Zaidi G, Rawool S, Malonia SK (2023) Next-generation sequencing technology: current trends and advancements. Biology 12(7):997. https://doi.org/10.3390/biology12070997
Schwengers O, Jelonek L, Dieckmann MA, Beyvers S, Blom J, Goesmann A (2021) Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb Genom 7(11):000685. https://doi.org/10.1099/mgen.0.000685
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068. https://doi.org/10.1093/bioinformatics/btu153
Sevgi E, Yılmaz OY, Çobanoğlu Özyiğitoğlu G, Tecimen HB, Sevgi O (2019) Factors influencing epiphytic lichen species distribution in a managed Mediterranean Pinus nigra Arnold Forest. Diversity 11(4):59. https://doi.org/10.3390/d11040059
Sharma PK, Balkwill DL, Frenkel A, Vairavamurthy MA (2000) A new Klebsiella planticola strain (Cd-1) grows anaerobically at high cadmium concentrations and precipitates cadmium sulfide. Appl Environ Microbiol 66(7):3083–3087. https://doi.org/10.1128/aem.66.7.3083-3087.2000
Sher S, Rehman A (2021) Proteomics and transcriptomic analysis of Micrococcus luteus strain AS2 under arsenite stress and its potential role in arsenic removal. Curr Res Microb Sci 2: 100020. https://doi.org/10.1016/j.crmicr.2021.100020 Sidhu GS (2016) Heavy metal toxicity in soils: sources, remediation technologies and challenges. Adv. Plants Agric Res 5(1): 445-6. https://doi.org/10.15406/apar.2016.05.00166
Shi Z, Yu X, Duan J, Guo W (2023) The complete genome sequence of Pseudomonas chengduensis BC1815 for genome mining of PET degrading enzymes. Mar Genom 67:101008. https://doi.org/10.1016/j.margen.2022.101008
Singh OV, Nagaraj NS (2006) Transcriptomics, proteomics and interactomics: unique approaches to track the insights of bioremediation. Brief Funct Genomics 4(4):355–362. https://doi.org/10.1093/bfgp/eli006
Sinha RK, Krishnan KP, Kurian PJ (2021) Complete genome sequence and comparative genome analysis of Alcanivorax sp. IO_7, a marine alkane-degrading bacterium isolated from hydrothermally-influenced deep seawater of southwest Indian ridge. Genomics 113(1):884–891. https://doi.org/10.1016/j.ygeno.2020.10.020
Slyemi D, Moinier D, Brochier-Armanet C, Bonnefoy V, Johnson DB (2011) Characteristics of a phylogenetically ambiguous, arsenic-oxidizing Thiomonas sp., Thiomonas arsenitoxydans strain 3As T sp. nov. Arch Microbiol 193:439–449. https://doi.org/10.1007/s00203-011-0684-y
Smirnova GF (2005) Distribution of bacteria resistant to oxygen-containing anions-xenobiotics. Mikrobiol Z 67:11–18
Sodhi KK, Singh CK, Kumar M, Singh DK (2023) Whole-genome sequencing of Alcaligenes sp. strain MMA: insight into the antibiotic and heavy metal resistant genes. Front Pharmacol 4:1144561. https://doi.org/10.3389/fphar.2023.1144561
Souza SS, Turcotte MR, Li J, Zhang X, Wolfe KL, Gao F, Andam CP (2022) Population analysis of heavy metal and biocide resistance genes in Salmonella enterica from human clinical cases in New Hampshire, United States. Front Microbiol 13:983083. https://doi.org/10.3389/fmicb.2022.983083
Sugawara M, Epstein B, Badgley BD, Unno T, Xu L, Reese J, Sadowsky MJ (2013) Comparative genomics of the core and accessory genomes of 48 Sinorhizobium strains comprising five genospecies. Genome Biol 14:1–20. https://doi.org/10.1186/gb-2013-14-2-r17
Summers AO (2002) Generally overlooked fundamentals of bacterial genetics and ecology. Clin Infect Dis 34:S85–S92. https://doi.org/10.1086/340245
Sun J, Zhou J, Wang Z, He W, Zhang D, Tong Q, Su X (2015) Multi-omics based changes in response to cadmium toxicity in Bacillus licheniformis a. RSC Adv 5(10):7330–7339. https://doi.org/10.1039/c4ra15280h
Sydor AM, Zamble DB (2013) Nickel metallomics: general themes guiding nickel homeostasis. In: Metallomics and the Cell, Singapore: Springer pp. 375–416. https://doi.org/10.1007/978-94-007-5561-1_11
Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Annika GL, Fang T, Doncheva NT, Pyysalo S, Bork P, Jensen LJ, von Mering C (2023) The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 51(D1):D638–D646. https://doi.org/10.1093/nar/gkac1000
Tayang A, Songachan LS (2021) Microbial bioremediation of heavy metals. Curr Sci 120(6):00113891. https://doi.org/10.18520/cs/v120/i6/1013-1025
Tellez-Plaza M, Navas-Acien A, Menke A, Crainiceanu CM, Pastor-Barriuso R, Guallar E (2012) Cadmium exposure and all-cause and cardiovascular mortality in the US general population. Environ Health Perspect 120(7):1017–1022. https://doi.org/10.1289/ehp.1104352
Tripathi AK, Saxena P, Thakur P, Rauniyar S, Samanta D, Gopalakrishnan V, Sani RK (2022) Transcriptomics and functional analysis of copper stress response in the sulfate-reducing bacterium Desulfovibrio alaskensis G20. Int J Mol Sci 23(3):1396. https://doi.org/10.3390/ijms23031396
Turner JS, Robinson NJ, Gupta A (1995) Construction of Zn2+/Cd2+-tolerant cynadobacteria with a modified metallothionein divergon: further analysis of the function and regulation of smt. J Ind Microbiol 14:259–264. https://doi.org/10.1007/bf01569937
UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res 43(D1). https://doi.org/10.1093/nar/gku989. D204-D212
Utturkar SM, Bollmann A, Brzoska RM, Klingeman DM, Epstein SE, Palumbo AV, Brown SD (2013) Draft genome sequence for Caulobacter sp. strain OR37, a bacterium tolerant to heavy metals. Genome Announc 1(3):10–1128. https://doi.org/10.1128/genomea.00322-13
Valasatava Y, Rosato A, Banci L, Andreini C (2016) MetalPredator: a web server to predict iron–sulfur cluster binding proteomes. Bioinformatics 32(18):2850–2852. https://doi.org/10.1093/bioinformatics/btw238
van der Lelie D, Springael D, Römling U, Ahmed N, Mergeay M (1999) Identification of a gene cluster, czr, involved in cadmium and zinc resistance in Pseudomonas aeruginosa. Gene 238(2):417–425. https://doi.org/10.1016/s0378-1119(99)00349-2
von Rozycki T, Nies DH (2009) Cupriavidus metallidurans: evolution of a metal-resistant bacterium. Antonie Leeuwenhoek 96:115–139. https://doi.org/10.1007/s10482-008-9284-5
Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Sobral BW (2014) PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42(D1):D581–D. https://doi.org/10.1093/nar/gkt1099
Wei W, Xia X, Wozniak M, Fan X, Damaševičius R, Li Y (2019) Multi-sink distributed power control algorithm for cyber-physical-systems in coal mine tunnels. Comput Netw 161:210–219. https://doi.org/10.1016/j.comnet.2019.04.017
Xia X, Li J, Zhou Z, Wang D, Huang J, Wang G (2018) High-quality-draft genome sequence of the multiple heavy metal resistant bacterium Pseudaminobacter manganicus JH-7T. Stand in Genomic Sci 13 (29) 1–8.https://doi.org/10.1186/s40793-018-0330-2
Xie Z, Wang D, Ben Fekih I, Yu Y, Li Y, Alwathnani H, Herzberg M, Rensing C (2023) Whole genome sequence analysis of Cupriavidus necator C39, a multiple heavy metal(loid) and antibiotic resistant bacterium isolated from a gold/copper mine. Microorganisms 11:1518. https://doi.org/10.3390/microorganisms11061518
Zahid N, Zulfiqar S, Shakoori AR (2012) Functional analysis of cus operon promoter of Klebsiella pneumoniae using E. coli lacZ assay. Gene 495(1): 81 – 8. https://doi.org/10.1016/j.gene.2011.12.040
Zaynab M, Al-Yahyai R, Ameen A, Sharif Y, Ali L, Fatima M, Khan KA, Li S (2022) Health and environmental effects of heavy metals. J King Saud Univ Sci 34(1):101653. https://doi.org/10.1016/j.jksus.2021.101653
Zefferino R, Piccoli C, Ricciardi N, Scrima R, Capitanio N (2017) Possible mechanisms of mercury toxicity and cancer promotion: Involvement of gap junction intercellular communications and inflammatory cytokines. Oxid. Med. Cell Longev. 2017: 1–6. https://doi.org/10.1155/2017/7028583
Zhang J (2003) Evolution by gene duplication: an update. Trends Ecol Evol 18(6):292–298. https://doi.org/10.1016/s0169-5347(03)00033-8
Ziller A, Fraissinet-Tachet L (2018) Metallothionein diversity and distribution in the tree of life: a multifunctional protein. Metallomics 10(11):1549–1559. https://doi.org/10.1039/c8mt00165k
Acknowledgements
The authors gratefully acknowledge the Deanship of Scientific Research, Vice Presidency for Graduate Studies and Scientific Research, King Faisal University, Saudi Arabia (GRANT 5872), for supporting this research work.
Author information
Authors and Affiliations
Contributions
A.A.H., H.S.E., O.A., B.A., A.R.H., A.A.R., H.H.A., A.M., N.I.E., A.A.A. wrote the main manuscript text and A.A.H, A.R.H. prepared Figs. 1-4. All authors reviewed the manuscript.”
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest related to this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Halema, A.A., El-Beltagi, H.S., Al-Dossary, O. et al. Omics technology draws a comprehensive heavy metal resistance strategy in bacteria. World J Microbiol Biotechnol 40, 193 (2024). https://doi.org/10.1007/s11274-024-04005-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-024-04005-y