Key message
A candidate gene, designate PpRPH, in the D locus was identified to control fruit acidity in peach.
Abstract
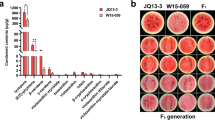
Fruit acidity has a strong impact on organoleptic quality of fruit. Peach fruit acidity is controlled by a large-effect D locus on chromosome 5. In this study, the D locus was mapped to a 509-kb interval, with two markers, 5dC720 and 5C1019, co-segregating with the non-acid/acid trait of peach fruit. Within this interval, a candidate gene encoding a putative small protein, designated PpRPH, showed a consistency between gene expression and fruit acidity, with up- and down-regulation in non-acidic and acidic fruits, respectively. Transient ectopic expression of PpRPH in tobacco leaves caused an increase of pH by approximately 40% compared to the control transformed with empty vector. Whereas, the concentrations of citrate and malate decreased significantly by 22% and 37%, respectively, with respect to the empty vector control. All these results suggest that PpRPH is a strong candidate gene of the D locus. These findings contribute to our overall understanding of the complex mechanism underlying fruit acidity in peach as well as that in other fruit crops.







Similar content being viewed by others
Data Availability
All data associated with this article can be found in the online version.
References
Arús P, Verde I, Sosinski B, Zhebentyayeva T, Abbott AG (2012) The peach genome. Tree Genet Genomes 8:531–547
Bae H, Yun SK, Jun JH, Yoon IK, Nam EY, Kwon JH (2014) Assessment of organic acid and sugar composition in apricot, plumcot, plum, and peach during fruit development. J Appl Bot Food Qual 87:24–29
Bai Y, Dougherty L, Li M, Fazio G, Cheng L, Xu K (2012) A natural mutation-led truncation in one of the two aluminum-activated malate transporter-like genes at the Ma locus is associated with low fruit acidity in apple. Mol Genet Genomics 287:663–678
Bielenberg DG, Wang YE, Li Z, Zhebentyayeva T, Fan S, Reighard GL, Scorza R, Abbott AG (2008) Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet Genomes 4:495–507
Boudehri K, Bendahmane A, Cardinet G, Troadec C, Moing A, Dirlewanger E (2009) Phenotypic and fine genetic characterization of the D locus controlling fruit acidity in peach. BMC Plant Biol 19:59
Butelli E, Licciardello C, Ramadugu C, Durand-Hulak M, Celant A, Reforgiato Recupero G, Froelicher Y, Martin C (2019) Noemi controls production of flavonoid pigments and fruit acidity and illustrates the domestication routes of modern citrus varieties. Curr Biol 29:158–164
Cantín CM, Moreno MA, Gogorcena Y (2009) Evaluation of the antioxidant capacity, phenolic compounds, and vitamin C content of different peach and nectarine [Prunus persica (L.) batsch] breeding progenies. J Agric Food Chem 57:4586–4592
Cao K, Zheng Z, Wang L, Liu X, Zhu G, Fang W et al (2014) Comparative population genomics reveals the domestication history of the peach, Prunus persica, and human influences on perennial fruit crops. Genome Biol 15:415
Cao K, Zhou Z, Wang Q, Guo J, Zhao P, Zhu G et al (2016) Genome-wide association study of 12 agronomic traits in peach. Nat Commun 11:13246
Cevallos-Casals BA, Byrne D, Okie WR, Cisneros-Zevallos L (2006) Selecting new peach and plum genotypes rich in phenolic compounds and enhanced functional properties. Food Chem 96:273–280
Cohen S, Itkin M, Yeselson Y, Tzuri G, Portnoy V, Harel-Baja R et al (2014) The PH gene determines fruit acidity and contributes to the evolution of sweet melons. Nat Commun 5:4026
Dirlewanger E, Moing A, Rothan C, Svanella L, Pronier V, Guye A et al (1999) Mapping QTL controlling fruit quality in peach (Prunus persica (L.) Batsch). Theor Appl Genet 98:18–31
Dirlewanger E, Graziano E, Joobeur T, Garriga-Caldere F, Cosson P, Howad W et al (2004) Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proc Natl Acad Sci USA 101:9891–9896
Dirlewanger E, Cosson P, Renaud C, Monet R, Poëssel JL, Moing A (2006) New detection of QTLs controlling major fruit quality components in peach. Acta Hortic 713:65–72
Desnoues E, Baldazzi V, Génard M, Mauroux JB, Lambert P, Confolent C, Quilot-Turion B (2016) Dynamic QTLs for sugars and enzyme activities provide an overview of genetic control of sugar metabolism during peach fruit development. J Exp Bot 67:3419–3431
Eduardo I, Pacheco I, Chietera G, Bassi D, Pozzi C, Vecchietti A et al (2011) QTL analysis of fruit quality traits in two peach intraspecific populations and importance of maturity date pleiotropic effect. Tree Genet Genomes 7:323–335
Eduardo I, López-Girona E, BatlIe I, Reig G, Iglesias I, Howad W et al (2014) Development of diagnostic markers for selection of the subacid trait in peach. Tree Genet Genomes 10:1695–1709
Etienne C, Rothan C, Moing A, Plomion C, Bodénès C, Svanella-Dumas L et al (2002) Candidate genes and QTLs for sugar and organic acid content in peach [ Prunus persica (L.) Batsch]. Theor Appl Genet 105:145–159
Hernández Mora JR, Micheletti D, Bink M, Van de Weg E, Cantín C, Nazzicari N et al (2017) Integrated QTL detection for key breeding traits in multiple peach progenies. BMC Genom 18:404
Howad W, Yamamoto T, Dirlewanger E, Testolin R, Cosson P, Cipriani G, Monforte AJ, Georgi L, Abbott AG, Arús P (2005) Mapping with a few plants: using selective mapping for microsatellite saturation of the Prunus reference map. Genetics 171:1305–1309
International Peach Genome Initiative (2013) The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 45:487–494
Jia D, Shen F, Wang Y, Wu T, Xu X, Zhang X, Han Z (2018) Apple fruit acidity is genetically diversified by natural variations in three hierarchical epistatic genes: MdSAUR37, MdPP2CH and MdALMTII. Plant J 95:427–443
Lambert P, Campoy JA, Pacheco I, Mauroux JB, Linge C, Micheletti D et al (2016) Identifying SNP markers tightly associated with six major genes in peach [Prunus persica (L.) Batsch] using a high-density SNP array with an objective of marker-assisted selection (MAS). Tree Genet Genomes 12:121
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li C, Dougherty L, Coluccio AE, Meng D, El-Sharkawy I, Borejsza-Wysocka E, Liang D, Piñeros MA, Xu K, Cheng L (2020) Apple ALMT9 Requires a Conserved C-Terminal Domain for Malate Transport Underlying Fruit Acidity. Plant Physiol 182:992–1006
Ma B, Liao L, Zheng H, Chen J, Wu B, Ogutu C, Li SH, Korban SS, Han Y (2015) Genes encoding aluminum-activated malate transporter II and their association with fruit acidity in apple. Plant Genome 8:3
Ma BQ, Zhao S, Wu BH, Wang D, Peng Q, Owiti A, Fang T, Liao L, Ogutu C, Korban SS, Li S, Han Y (2016) Construction of a high density linkage map and its application in the identification of QTLs for soluble sugar and organic acid components in apple. Tree Genet Genomes 12:1
Ma B, Liao L, Fang T, Peng Q, Ogutu C, Zhou H et al (2019) A Ma10 gene encoding P-type ATPase is involved in fruit organic acid accumulation in apple. Plant Biotechnol J 17:674–686
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Micheletti D, Dettori MT, Micali S, Aramini V, Pacheco I, Da Silva Linge C et al (2015) Whole-Genome Analysis of Diversity and SNP-Major Gene Association in Peach Germplasm. PLoS One 10:e0136803
Morvai M, Molnár-Perl I (1992) Simultaneous gas chromatographic quantitation of sugars and acids in citrus fruits, pears, bananas, grapes, apples and tomatoes. Chromatographia 34:502–504
Oh E, Seo PJ, Kim J (2018) Signaling peptides and receptors coordinating plant root development. Trends Plant Sci 23:337–351
Peng Q, Wang L, Ogutu C, Liu J, Liu L, Mollah MDA, Han Y (2020) Functional analysis reveals the regulatory role of PpTST1 encoding tonoplast sugar transporter in sugar accumulation of peach fruit. Int J Mol Sci 21:1112
Strazzer P, Spelt CE, Li S, Bliek M, Federici CT, Roose ML, Koes R, Quattrocchio FM (2019) Hyperacidification of Citrus fruits by a vacuolar proton-pumping P-ATPase complex. Nat Commun 10:744
Sugio T, Satoh J, Matsuura H, Shinmyo A, Kato K (2008) The 5’-untranslated region of the Oryza sativa alcohol dehydrogenase gene functions as a translational enhancer in monocotyledonous plant cells. J Biosci Bioeng 105(3):300–302
Van Ooijen J (2006) JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen
Verde I, Bassil N, Scalabrin S, Gilmore B, Lawley CT, Gasic K, Micheletti D, Rosyara UR, Cattonaro F, Vendramin E, Main D, Aramini V, Blas AL, Mockler TC, Bryant DW, Wilhelm L, Troggio M, Sosinski B, Aranzana MJ, Arús P, Iezzoni A, Morgante M, Peace C (2012) Development and evaluation of a 9K SNP array for peach by internationally coordinated SNP detection and validation in breeding germplasm. PLoS One 7:e35668
Vizzotto M, Cisneros-Zevallos L, Byrne DH, Ramming DW, Okie WR (2007) Large variation found in the phytochemical and antioxidant activity of peach and plum germplasm. J Amer Soc HoRT SCI 132:334–340
Wang L, Zhao S, Gu C, Zhou Y, Zhou H, Ma J, Cheng J, Han Y (2013) Deep RNA-Seq uncovers the peach transcriptome landscape. Plant Mol Biol 83:365–377
Ye J, Wang X, Hu T, Zhang F, Wang B, Li C et al (2017) An inDel in the promoter of Al-ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell 29:2249–2268
Zeballos JL, Abidi W, Giménez R, Monforte AJ, Moreno MA, Gogorcena Y (2016) Mapping QTLs associated with fruit quality traits in peach [Prunus persica (L.) Batsch] using SNP maps. Tree Genet Genomes 12:37
Zhang Q, Ma B, Li H, Chang Y, Han Y, Li J, Wei G, Zhao S, Khan MA, Zhou Y, Gu C, Zhang X, Han Z, Korban SS, Li S, Han Y (2012) Identification, characterization, and utilization of genome-wide simple sequence repeats to identify a QTL for acidity in apple. BMC Genom 13:537
Zhao G, Lian Q, Zhang Z, Fu Q, He Y, Ma S et al (2019) A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat Genet 51:1607–1615
Acknowledgements
We would like to thank Dr. Lei Gao for his help in analysis of resequencing data.
Funding
This work was supported by funds received from the National Key R&D Program of China (2019YFD1000200), the National Natural Science Foundation of China (31672134) and the China Agriculture Research System (CARS-30).
Author information
Authors and Affiliations
Contributions
W.L. conducted most experiments of this study and wrote the manuscript. JX performed the transcriptomic analysis. ZL, FW, HZ and HH prepared the experimental materials. L.Y. participated in analysis of genotyping data. Y.H. was overall project leader and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, L., Jiang, X., Zhao, L. et al. A candidate PpRPH gene of the D locus controlling fruit acidity in peach. Plant Mol Biol 105, 321–332 (2021). https://doi.org/10.1007/s11103-020-01089-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-01089-6