Abstract
Key message
This study precisely mapped and validated a quantitative trait locus (QTL) located on chromosome 4B for flag leaf angle in wheat.
Abstract
Flag leaf angle (FLANG) is closely related to crop architecture and yield. We previously identified the quantitative trait locus (QTL) QFLANG-4B for FLANG on chromosome 4B, located within a 14-cM interval flanked by the markers Xbarc20 and Xzyh357, using a mapping population of recombinant inbred lines (RILs) derived from a cross between Nongda3331 (ND3331) and Zang1817. In this study, we fine-mapped QFLANG-4B and validated its associated genetic effect. We developed a BC3F3 population using ND3331 as the recurrent parent through marker-assisted selection, as well as near-isogenic lines (NILs) by selfing BC3F3 plants carrying different heterozygous segments for the QFLANG-4B region. We obtained eight recombinant types for QFLANG-4B, narrowing its location down to a 5.3-Mb region. This region contained 76 predicted genes, 7 of which we considered to be likely candidate genes for QFLANG-4B. Marker and phenotypic analyses of individual plants from the secondary mapping populations and their progeny revealed that the FLANG of the ND3331 allele is significantly higher than that of the Zang1817 allele in multiple environments. These results not only provide a basis for the map-based cloning of QFLANG-4B, but also indicate that QFLANG-4B has great potential for marker-assisted selection in wheat breeding programs designed to improve plant architecture and yield.





Similar content being viewed by others
Data availability
RNA sequencing data are available at the Sequence Read Archive (SRA) under accession no. PRJNA1070335.
References
Cai J, Zhang M, Guo LB, Li XM, Bao JS, Ma LY (2015) QTLs for rice flag leaf traits in doubled haploid populations in different environments. Genet Mol Res 14:6786–6795
Cao Y, Zhong Z, Wang H, Shen R (2022) Leaf angle: a target of genetic improvement in cereal crops tailored for high-density planting. Plant Biotechnol J 20:426–436
Chang S, Chen Q, Yang T, Li B, Xin M, Su Z, Du J, Guo W, Hu Z, Liu J, Peng H, Ni Z, Sun Q, Yao Y (2022) Pinb-D1p is an elite allele for improving end-use quality in wheat (Triticum aestivum L.). Theor Appl Genet 135:4469–4481
Chen YM, Song WJ, Xie XM, Wang ZH, Guan PF, Peng HR, Jiao YN, Ni ZF, Sun QX, Guo WL (2020) A Collinearity-Incorporating Homology Inference Strategy for Connecting Emerging Assemblies in the Triticeae Tribe as a Pilot Practice in the Plant Pangenomic Era. Mol Plant 13:1694–1708
Chen S, Liu F, Wu W, Jiang Y, Zhan K (2021) A SNP-based GWAS and functional haplotype-based GWAS of flag leaf-related traits and their influence on the yield of bread wheat (Triticum aestivum L.). Theor Appl Genet 134:3895–3909
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Donald CM (1968) The breeding of crop ideotypes. Euphytica 17:385–403
Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, Uozu S, Kitano H, Ashikari M, Matsuoka M (2002) Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J 32:495–508
Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M (2003) A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15:2900–2910
Kim SH, Yoon J, Kim H, Lee SJ, Kim T, Kang K, Paek NC (2023) OsMYB7 determines leaf angle at the late developmental stage of lamina joints in rice. Front Plant Sci 14:1167202
Kong D, Wang B, Wang H (2020) UPA2 and ZmRAVL1: Promising targets of genetic improvement of maize plant architecture. J Integr Plant Biol 62:394–397
Ku LX, Zhao WM, Zhang J, Wu LC, Wang CL, Wang PA, Zhang WQ, Chen YH (2010) Quantitative trait loci mapping of leaf angle and leaf orientation value in maize (Zea mays L.). Theor Appl Genet 121:951–959
Lee J, Park J-J, Kim SL, Yim J, An G (2007) Mutations in the rice liguleless gene result in a complete loss of the auricle, ligule, and laminar joint. Plant Mol Biol 65:487–499
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li X, Wu P, Lu Y, Guo S, Zhong Z, Shen R, Xie Q (2020) Synergistic interaction of phytohormones in determining leaf angle in crops. Int J Mol Sci 21(14):5052
Li Q, Xu F, Chen Z, Teng Z, Sun K, Li X, Yu J, Zhang G, Liang Y, Huang X, Du L, Qian Y, Wang Y, Chu C, Tang J (2021) Synergistic interplay of ABA and BR signal in regulating plant growth and adaptation. Nature Plants 7:1108–1118
Liu G, Jia LJ, Lu LH, Qin DD, Zhang JP, Guan PF, Ni ZF, Yao YY, Sun QX, Peng HR (2014) Mapping QTLs of yield-related traits using RIL population derived from common wheat and Tibetan semi-wild wheat. Theor Appl Genet 127:2415–2432
Liu K, Xu H, Liu G, Guan P, Zhou X, Peng H, Yao Y, Ni Z, Sun Q, Du J (2018) QTL mapping of flag leaf-related traits in wheat (Triticum aestivum L.). Theor Appl Genet 131:839–849
Liu K, Cao J, Yu K, Liu X, Gao Y, Chen Q, Zhang W, Peng H, Du J, Xin M, Hu Z, Guo W, Rossi V, Ni Z, Sun Q, Yao Y (2019) Wheat TaSPL8 modulates leaf angle through Auxin and brassinosteroid signaling. Plant Physiol 181:179–194
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15
Luo X, Zheng J, Huang R, Huang Y, Wang H, Jiang L, Fang X (2016) Phytohormones signaling and crosstalk regulating leaf angle in rice. Plant Cell Rep 35:2423–2433
Ma J, Tu Y, Zhu J, Luo W, Liu H, Li C, Li S, Liu J, Ding P, Habib A, Mu Y, Tang H, Liu Y, Jiang Q, Chen G, Wang J, Li W, Pu Z, Zheng Y, Wei Y, Kang H, Chen G, Lan X (2020) Flag leaf size and posture of bread wheat: genetic dissection, QTL validation and their relationships with yield-related traits. Theor Appl Genet 133:297–315
Mantilla-Perez MB, Salas Fernandez MG (2017) Differential manipulation of leaf angle throughout the canopy: current status and prospects. J Exp Bot 68:5699–5717
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Moreno MA, Harper LC, Krueger RW, Dellaporta SL, Freeling M (1997) liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev 11:616–628
Qu R, Zhang P, Liu Q, Wang Y, Guo W, Du Z, Li X, Yang L, Yan S, Gu X (2022) Genome-edited ATP BINDING CASSETTE B1 transporter SD8 knockouts show optimized rice architecture without yield penalty. Plant Commun 3:100347
Seo H, Kim SH, Lee BD, Lim JH, Lee SJ, An G, Paek NC (2020) The rice basic helix-loop-helix 79 (OsbHLH079) determines leaf angle and grain shape. Int J Mol Sci 21(6):2090
Sharma SN, Sain RS, Sharma RK (2004) The genetic control of flag leaf length in normal and late sown durum wheat. J Agric Sci 141:323–331
Silva Lda CWS, Zeng ZB (2012) Composite interval mapping and multiple interval mapping: procedures and guidelines for using Windows QTL Cartographer. Methods Mol Biol 871:75–119
Song L, Liu J, Cao B, Liu B, Zhang X, Chen Z, Dong C, Liu X, Zhang Z, Wang W, Chai L, Liu J, Zhu J, Cui S, He F, Peng H, Hu Z, Su Z, Guo W, Xin M, Yao Y, Yan Y, Song Y, Bai G, Sun Q, Ni Z (2023) Reducing brassinosteroid signalling enhances grain yield in semi-dwarf wheat. Nature 617:118–124
Stam P (1993) Construction of integrated genetic linkage maps by means of a new computer package: Join Map. Plant J 3:739–744
Tanabe S, Ashikari M, Fujioka S, Takatsuto S, Yoshida S, Yano M, Yoshimura A, Kitano H, Matsuoka M, Fujisawa Y, Kato H, Iwasaki Y (2005) A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf Mutant, dwarf11, with reduced seed length. Plant Cell 17:776–790
Tian J, Wang C, Xia J, Wu L, Xu G, Wu W, Li D, Qin W, Han X, Chen Q, Jin W, Tian F (2019) Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science (New York, NY) 365:658–664
Tong H, Liu L, Jin Y, Du L, Yin Y, Qian Q, Zhu L, Chu C (2012) Dwarf and low-tillering acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24:2562–2577
Tong H, Xiao Y, Liu D, Gao S, Liu L, Yin Y, Jin Y, Qian Q, Chu C (2014) Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26:4376–4393
Tu Y, Liu H, Liu J, Tang H, Mu Y, Deng M, Jiang Q, Liu Y, Chen G, Wang J, Qi P, Pu Z, Chen G, Peng Y, Jiang Y, Xu Q, Kang H, Lan X, Wei Y, Zheng Y, Ma J (2021) QTL mapping and validation of bread wheat flag leaf morphology across multiple environments in different genetic backgrounds. Theor Appl Genet 134:261–278
Walsh J, Waters CA, Freeling M (1998) The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes Dev 12:208–218
Wang B, Smith SM, Li J (2018) Genetic regulation of shoot architecture. Annu Rev Plant Biol 69:437–468
Wang K, Li MQ, Chang YP, Zhang B, Zhao QZ, Zhao WL (2020) The basic helix-loop-helix transcription factor OsBLR1 regulates leaf angle in rice via brassinosteroid signalling. Plant Mol Biol 102:589–602
Wright IJ, Reich PB, Westoby M, Ackerly DD, Baruch Z, Bongers F, Cavender-Bares J, Chapin T, Cornelissen JHC, Diemer M, Flexas J, Garnier E, Groom PK, Gulias J, Hikosaka K, Lamont BB, Lee T, Lee W, Lusk C, Midgley JJ, Navas M-L, Niinemets Ü, Oleksyn J, Osada N, Poorter H, Poot P, Prior L, Pyankov VI, Roumet C, Thomas SC, Tjoelker MG, Veneklaas EJ, Villar R (2004) The worldwide leaf economics spectrum. Nature 428:821–827
Yang X, Li R, Jablonski A, Stovall A, Kim J, Yi K, Ma Y, Beverly D, Phillips R, Novick K, Xu X, Lerdau M (2023) Leaf angle as a leaf and canopy trait: Rejuvenating its role in ecology with new technology. Ecol Lett 26:1005–1020
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant no. 32125030, U22A20478), Pinduoduo-China Agricultural University Research Fund (PC2023A01003), Major Program of National Agricultural Science and Technology of China (NK20220607).
Funding
Fund was provided by National Natural Science Foundation of China, Grant No. 32125030, Yingyin Yao, Pinduoduo-China Agricultural University Research Fund, PC2023A01003, Yingyin Yao, Major Program of National Fund of Philosophy and Social Science of China, NK20220607, Jinkun Du.
Author information
Authors and Affiliations
Contributions
JD conceived the project; WZ performed the experiments; XC, KY, SC, and XZ provided technological assistance; ML and LW performed bioinformatics analysis; MX, ZH, JL, HP, ZN, QS, and YY provided theoretical contributions to the project; WZ and JD contributed to writing and revision of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Philomin Juliana.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
122_2024_4629_MOESM1_ESM.jpg
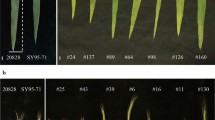
Phenotypic variations in the flag leaves of ND3331, Zang1817, and R53 during the flowering stage. Flag leaf angle (FLANG) is the angle between the stem immediately under the spike and the fag leaf midrib. Scale bars, 5 cm. (JPG 149 kb)
122_2024_4629_MOESM2_ESM.jpg
Fine-mapping of QFLANG-4B. (a) Genetic location of the QFLANG-4B mapping interval on chromosome 4B. (b) Diagram of the 14 recombinant genotypes from the BC3F3 population in the ND3331 background. Black, homozygous for ND3331; gray, heterozygous; white, homozygous Zang1817. (JPG 299 kb)
122_2024_4629_MOESM3_ESM.jpg
Effect of the genotype at the QFLANG-4B region in two bulks of from different NIL pairs on mean FLANG values. ***, P < 0.001 as determined by a two-tailed Student’s t test. (JPG 808 kb)
122_2024_4629_MOESM4_ESM.jpg
Colinearity analysis of the QFLANG-4B mapping interval between the Zang1817 and Chinese Spring genomes. The reference genome version IWGSC RefSeq v1.1 was used for Chinese Spring. The genes and lines of homologous gene pairs are grouped by their strength of relationship. Color priority: RBH (Reciprocal Best Hit) > SBH (Single-side Best Hit) > 1-to-many (all putative homologous genes). All homologous lines are grouped by score: 0–50, 50–70, and 70–100. (JPG 680 kb)
122_2024_4629_MOESM5_ESM.jpg
Distribution of single nucleotide polymorphisms (SNPs) on chromosomes. Sliding window method was used for reducing noise. (JPG 1147 kb)
122_2024_4629_MOESM6_ESM.jpg
Analysis of genes expressed at very low levels (TPM ≤ 1) (a) and with similar relative expression levels (b) in the two bulks of homozygous ND3331 and Zang1817 within the QFLANG-4B mapping interval at the heading stage. (JPG 1394 kb)
122_2024_4629_MOESM7_ESM.jpg
(a) Sequence variations of “r-e-z haploblock” between the parents ND3331 and Zang1817. Comparison of the genomic sequences between ND3331 and Zang1817 within the QTL region revealed a large fragment deletion (~500 kb) spanning three high confident genes including ZnF-B, EamA-B and Rht-B1a in ND3331. (b) Heatmap representing the transcriptional changes in TaSPL8’s three homoeoalleles on chromosome 2A, 2B and 2D in the bulk of homozygous Zang1817 for QFlANG-4B relative to the bulk of homozygous ND3331. Heat map illustrating the FPKM-based expression patterns of these genes. For each gene, the average FPKM values of three biological replicates for each sample were normalized using R and reported in the heatmap. (JPG 381 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, W., Chen, X., Yang, K. et al. Fine-mapping and validation of the major quantitative trait locus QFlANG-4B for flag leaf angle in wheat. Theor Appl Genet 137, 121 (2024). https://doi.org/10.1007/s00122-024-04629-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-024-04629-6