Abstract
Climate change poses a significant threat to the global ecosystem, prompting plants to use various adaptive mechanisms via molecular switches to combat biotic and abiotic stress factors. These switches activate stress-induced pathways by altering their configuration between stable states. In this review, we investigated the regulation of molecular switches in different plant species in response to stress, including the stress-regulated response of multiple switches in Arabidopsis thaliana. We also discussed techniques for developing stress-resilient crops using molecular switches through advanced biotechnological tools. The literature search, conducted using databases such as PubMed, Google Scholar, Web of Science, and SCOPUS, utilized keywords such as molecular switch, plant adaptation, biotic and abiotic stresses, transcription factors, Arabidopsis thaliana, and crop improvement. Recent studies have shown that a single molecular switch can regulate multiple stress networks, and multiple switches can regulate a single stress condition. This multifactorial understanding provides clarity to the switch regulatory network and highlights the interrelationships of different molecular switches. Advanced breeding techniques, along with genomic and biotechnological tools, have paved the way for further research on molecular switches in crop improvement. The use of synthetic biology in molecular switches will lead to a better understanding of plant stress biology and potentially bring forth a new era of stress-resilient, climate-smart crops worldwide.






Similar content being viewed by others
Data Availability
Data sharing does not apply to this article as no datasets were generated or analyzed during the current study.
References
TNAU Portal. http://www.agritech.tnau.ac.in/agriculture/agri_agrometeorology_temp.htm
Liu X, Zhou Y, Xiao J et al (2018) Effects of chilling on the structure, function and development of chloroplasts. Front Plant Sci 9:1715. https://doi.org/10.3389/fpls.2018.01715
Srikant T, Drost HG (2021) How stress facilitates phenotypic innovation through epigenetic diversity. Front Plant Sci 11:606800. https://doi.org/10.3389/fpls.2020.606800
Park HJ, Kim WY, Yun DJ (2016) A new insight of salt stress signaling in plant. Mol Cells 39(6):447–459. https://doi.org/10.14348/molcells.2016.0083
Liang X, Zhang L, Natarajan SK et al (2013) Proline mechanisms of stress survival. Antioxid Redox Signal 19(9):998–1011. https://doi.org/10.1089/ars.2012.5074
Berken A (2006) ROPs in the spotlight of plant signal transduction. Cell Mol Life Sci 63(21):2446–2459. https://doi.org/10.1007/s00018-006-6197-1
Fang XZ, Fang SQ, Ye ZQ et al (2021) NRT1.1 dual-affinity nitrate transport/signalling and its roles in plant abiotic stress resistance. Front Plant Sci 12:715694. https://doi.org/10.3389/fpls.2021.715694
Leng P, Zhao J (2020) Transcription factors as molecular switches to regulate drought adaptation in maize. Theor Appl Genet 133(5):1455–1465. https://doi.org/10.1007/s00122-019-03494-y
Maurya JP, Triozzi PM, Bhalerao RP et al (2018) Environmentally sensitive molecular switches drive poplar phenology. Front Plant Sci 9:1873. https://doi.org/10.3389/fpls.2018.01873
Joshi R, Wani SH, Singh B et al (2016) Transcription factors and plants response to drought stress: current understanding and future directions. Front Plant Sci 7:1029. https://doi.org/10.3389/fpls.2016.01029
Adem GD, Chen G, Shabala L et al (2020) GORK Channel: a Master switch of Plant Metabolism? Trends Plant Sci 25(5):434–445. https://doi.org/10.1016/j.tplants.2019.12.012
Duszyn M, Świeżawska B, Szmidt-Jaworska A et al (2019) Cyclic nucleotide gated channels (CNGCs) in plant signalling-current knowledge and perspectives. J Plant Physiol 241:153035. https://doi.org/10.1016/j.jplph.2019.153035
Del Carmen Martínez-Ballesta M, Moreno DA, Carvajal M (2013) The physiological importance of glucosinolates on plant response to abiotic stress in Brassica. Int J Mol Sci 14(6):11607–11625. https://doi.org/10.3390/ijms140611607
Perrone A, Martinelli F (2020) Plant stress biology in epigenomic era. Plant Sci 294:110376. https://doi.org/10.1016/j.plantsci.2019.110376
Khong G, Richaud F, Coudert Y et al (2008) Modulating rice stress tolerance by transcription factors. Biotechnol Genet Eng Rev 25:381–403. https://doi.org/10.5661/bger-25-381
Ren M, Wang Z, Xue M et al (2019) Constitutive expression of an A-5 subgroup member in the DREB transcription factor subfamily from Ammopiptanthus mongolicus enhanced abiotic stress tolerance and anthocyanin accumulation in transgenic Arabidopsis. PLoS ONE 14(10):e0224296. https://doi.org/10.1371/journal.pone.0224296
Mizoi J, Ohori T, Moriwaki T et al (2013) GmDREB2A;2, a canonical DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol 161(1):346–361. https://doi.org/10.1104/pp.112.204875
Qin QL, Liu JG, Zhang Z et al (2007b) Isolation, optimization, and functional analysis of the cDNA encoding transcription factor OsDREB1B in Oryza sativa L. Mol Breed 19(4):329–340. https://doi.org/10.1007/s10725-013-9866-8
Wang Q, Guan Y, Wu Y et al (2008) Overexpression of a rice OsDREB1F gene increases salt, drought and low temperature tolerance in both Arabidopsis and rice. Plant Mol Biol 67:589–602. https://doi.org/10.1007/s11103-008-9340-6
Du X, Li W, Sheng L et al (2018) Over-expression of chrysanthemum CmDREB6 enhanced tolerance of chrysanthemum to heat stress. BMC Plant Biol 18(1):1–10. https://doi.org/10.1186/s12870-018-1400-8
Morran S, Eini O, Pyvovarenko T et al (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J 9(2):230–249. https://doi.org/10.1111/j.1467-7652.2010.00547.x
Hong B, Ma C, Yang Y et al (2009) Over-expression of AtDREB1A in chrysanthemum enhances tolerance to heat stress. Plant Mol Biol 70(3):231–240. https://doi.org/10.1007/s11103-009-9468-z
Le Hir R, Castelain M, Chakraborti D et al (2017) AtbHLH68 transcription factor contributes to the regulation of ABA homeostasis and drought stress tolerance in Arabidopsis thaliana. Physiol Plant 160(3):312–327. https://doi.org/10.1111/ppl.12549
Liu Y, Ji X, Nie X et al (2015) Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol 207(3):692–709. https://doi.org/10.1111/nph.13387
Huang D, Dai W (2015) Molecular characterization of the basic helix-loop-helix (bHLH) genes that are differentially expressed and induced by iron Deficiency in Populus. Plant Cell Rep 34(7):1211–1224. https://doi.org/10.1007/s00299-015-1779-8
Chen HC, Hsieh-Feng V, Liao PC et al (2017) The function of OsbHLH068 is partially redundant with its homolog, AtbHLH112, in the regulation of the salt stress response but has opposite functions to control flowering in Arabidopsis. Plant Mol Biol 94(4–5):531–548. https://doi.org/10.1007/s11103-017-0624-6
Qiu Z, Wang X, Gao J et al (2016) The tomato Hoffman’s anthocyaninless gene encodes a bHLH transcription factor involved in anthocyanin biosynthesis that is developmentally regulated and induced by low temperatures. PLoS ONE 11(3):e0151067. https://doi.org/10.1371/journal.pone.0151067
Jia Z, Lian Y, Zhu Y et al (2009) Cloning and characterization of a putative transcription factor induced by abiotic stress in Zea mays. Afr J Biotechnol 8(24):6764–6771. https://doi.org/10.5897/AJB2009.000-9520
Gai WX, Ma X, Qiao YM et al (2020) Characterization of the bZIP transcription factor family in Pepper (Capsicum annuum L.): CabZIP25 positively modulates the Salt Tolerance. Front Plant Sci 11:139. https://doi.org/10.3389/fpls.2020.00139
Chen L, Song Y, Li S et al (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochimn Biophys Acta 1819(2):120–128. https://doi.org/10.1016/j.bbagrm.2011.09.002
Xue GP, Loveridge CW (2004) HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J 37(3):326–339. https://doi.org/10.1046/j.1365-313x.2003.01963.x
Xiang Y, Tang N, Du H et al (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148(4):1938–1952. https://doi.org/10.1104/pp.108.128199
Chang Y, Nguyen BH, Xie Y et al (2017) Co-overexpression of the constitutively active form of OsbZIP46 and ABA-Activated protein kinase SAPK6 improves Drought and temperature stress resistance in Rice. Front Plant Sci 8:1102. https://doi.org/10.3389/fpls.2017.01102
Lee HG, Seo PJ (2015) The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J 82(6):962–977. https://doi.org/10.1111/tpj.12866
El-Kereamy A, Bi YM, Ranathunge K et al (2012) The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE 7(12):e52030. https://doi.org/10.1371/journal.pone.0052030
Guo C, Yao L, You C et al (2016) MID1 plays an important role in response to drought stress during reproductive development. Plant J 88(2):280–293. https://doi.org/10.1111/tpj.13250
Liao C, Zheng Y, Guo Y (2017) MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol 216(1):163–177. https://doi.org/10.1111/nph.14679
Zhu N, Cheng S, Liu X et al (2015) The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci 236:146–156. https://doi.org/10.1016/j.plantsci.2015.03.023
Campos JF, Cara B, Pérez-Martín F et al (2016) The tomato mutant ars1 (altered response to salt stress 1) identifies an R1-type MYB transcription factor involved in stomatal closure under salt acclimation. Plant Biotechnol J 14(6):1345–1356. https://doi.org/10.1111/pbi.12498
Feng C, Andreasson E, Maslak A et al (2004) Arabidopsis MYB68 in development and responses to environmental cues. Plant Sci 167(5):1099–1107. https://doi.org/10.1016/j.plantsci.2004.06.014
Ebrahimian-Motlagh S, Ribone PA, Thirumalaikumar VP et al (2017) JUNGBRUNNEN1 confers Drought Tolerance downstream of the HD-Zip I transcription factor AtHB13. Front Plant Sci 8:2118. https://doi.org/10.3389/fpls.2017.02118
Song SY, Chen Y, Chen J et al (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234(2):331–345. https://doi.org/10.1007/s00425-011-1403-2
Guan Q, Yue X, Zeng H et al (2014) The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell 26(1):438–453. https://doi.org/10.1105/tpc.113.118927
He L, Shi X, Wang Y et al (2017) Arabidopsis ANAC069 binds to C[A/G]CG[T/G] sequences to negatively regulate salt and osmotic stress tolerance. Plant Mol Biol 93(4–5):369–387. https://doi.org/10.1007/s11103-016-0567-3
Hao YJ, Wei W, Song QX et al (2011) Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J 68(2):302–313. https://doi.org/10.1111/j.1365-313X.2011.04687.x
Hong Y, Zhang H, Huang L et al (2016) Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves Drought and Salt Tolerance in Rice. Front Plant Sci 7:4. https://doi.org/10.3389/fpls.2016.00004
Xue GP, Way HM, Richardson T et al (2011) Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 4(4):697–712. https://doi.org/10.1093/mp/ssr013
Li XD, Zhuang KY, Liu ZM et al (2016) Overexpression of a novel NAC-type tomato transcription factor, SlNAM1, enhances the chilling stress tolerance of transgenic Tobacco. J Plant Physiol 204:54–65. https://doi.org/10.1016/j.jplph.2016.06.024
Khan SA, Li MZ, Wang SM et al (2018) Revisiting the role of plant transcription factors in the battle against Abiotic stress. Int J Mol Sci 19(6):1634. https://doi.org/10.3390/ijms19061634
Li S, Zhou X, Chen L et al (2010b) Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol Cells 29(5):475–483. https://doi.org/10.1007/s10059-010-0059-2
Liu L, Zhang Z, Dong J et al (2016) Overexpression of MtWRKY76 increases both salt and drought tolerance in Medicago truncatula. Environ Exp Bot 123:50–58. https://doi.org/10.1016/j.envexpbot.2015.10.007
Li S, Fu Q, Chen L et al (2011) Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233(6):1237–1252. https://doi.org/10.1007/s00425-011-1375-2
Hichri I, Muhovski Y, Žižková E et al (2017) The Solanum lycopersicum WRKY3 transcription factor SlWRKY3 is involved in salt stress tolerance in tomato. Front Plant Sci 8:1343. https://doi.org/10.3389/fpls.2017.01343
Sun Y, Yu D (2015) Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep 34(8):1295–1306. https://doi.org/10.1007/s00299-015-1787-8
Li Y, Williams B, Dickman M (2017) Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG 7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY 29. New Phytol 214(2):695–705. https://doi.org/10.1111/nph.14388
Chen HC, Chien TC, Chen TY et al (2021) Overexpression of a novel ERF-X-Type transcription factor, OsERF106MZ, reduces shoot growth and tolerance to salinity stress in Rice. Rice (N Y) 14(1):82. https://doi.org/10.1186/s12284-021-00525-5
Li XW, Wang Y, Yan F et al (2016) Overexpression of soybean R2R3-MYB transcription factor, GmMYB12B2, and tolerance to UV radiation and salt stress in transgenic Arabidopsis. Genet Mol Res 15(2). https://doi.org/10.4238/gmr.15026573
Zhang Z, Hu X, Zhang Y et al (2016) Opposing control by transcription factors MYB61 and MYB3 increases freezing tolerance by relieving C-Repeat binding factor suppression. Plant Physiol 172(2):1306–1323. https://doi.org/10.1104/pp.16.00051
Ullah A, Sun H, Hakim et al (2018) A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol Plant 162(4):439–454. https://doi.org/10.1111/ppl.12651
Yang T, Hao L, Yao S et al (2016) TabHLH1, a bHLH-type transcription factor gene in wheat, improves plant tolerance to pi and N deprivation via regulation of nutrient transporter gene transcription and ROS homeostasis. Plant Physiol Biochem 104:99–113. https://doi.org/10.1016/j.plaphy.2016.03.023
Wang N, Cui Y, Liu Y et al (2013) Requirement and functional redundancy of ib subgroup bHLH proteins for iron Deficiency responses and uptake in Arabidopsis thaliana. Mol Plant 6(2):503–513. https://doi.org/10.1093/mp/sss089
Jha SK, Sharma M, Pandey GK (2016) Role of cyclic nucleotide gated channels in stress management in plants. Curr Genomics 17(4):315–329. https://doi.org/10.2174/1389202917666160331202125
Jian S, Luo J, Liao Q et al (2019) NRT1.1 regulates nitrate allocation and Cadmium Tolerance in Arabidopsis. Front Plant Sci 10:384. https://doi.org/10.3389/fpls.2019.00384
Andres J, Blomeier T, Zurbriggen MD (2019) Synthetic switches and Regulatory circuits in plants. Plant Physiol 179(3):862–884. https://doi.org/10.1104/pp.18.01362
Zhang P, Qian D, Luo C et al (2021) Arabidopsis ADF5 acts as a downstream target gene of CBFs in response to low-temperature stress. Front Cell Dev Biol 9:635533. https://doi.org/10.3389/fcell.2021.635533
Jiang Y, Peng D, Bai LP et al (2013) Molecular switch for cold acclimation -- anatomy of the cold-inducible promoter in plants. Biochem (Mosc) 78(4):342–354. https://doi.org/10.1134/S0006297913040032
Shi Y, Huang J, Sun T et al (2017) The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana. J Integr Plant Biol 59(2):118–133. https://doi.org/10.1111/jipb.12515
Horvath DP, McLarney BK, Thomashow MF (1993) Regulation of Arabidopsis thaliana L. (Heyn) cor78 in response to low temperature. Plant Physiol 103(4):1047–1453. https://doi.org/10.1104/pp.103.4.1047
Artus NN, Uemura M, Steponkus PL et al (1996) Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci U S A 93(23):13404–13409. https://doi.org/10.1073/pnas.93.23.13404
Gong Z, Lee H, Xiong L et al (2002) RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci U S A 99(17):11507–11512. https://doi.org/10.1073/pnas.172399299
Fowler SG, Cook D, Thomashow MF (2005) Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol 137(3):961–968. https://doi.org/10.1104/pp.104.058354
Charng YY, Liu HC, Liu NY et al (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143(1):251–262. https://doi.org/10.1104/pp.106.091322
Haider S, Iqbal J, Naseer S et al (2022) Unfolding molecular switches in plant heat stress resistance: a comprehensive review. Plant Cell Rep 41(3):775–798. https://doi.org/10.1007/s00299-021-02754-w
Eulgem T, Rushton PJ, Robatzek S et al (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5(5):199–206. https://doi.org/10.1016/s1360-1385(00)01600-9
Li S, Fu Q, Huang W, Yu D (2009) Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep 28(4):683–693. https://doi.org/10.1007/s00299-008-0666-y
Lee S, Lee HJ, Huh SU et al (2014) The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci 227:76–83. https://doi.org/10.1016/j.plantsci.2014.07.003
Schramm F, Larkindale J, Kiehlmann E et al (2008) A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J 53(2):264–274. https://doi.org/10.1111/j.1365-313X.2007.03334.x
Yoshida T, Sakuma Y, Todaka D et al (2008) Functional analysis of an Arabidopsis heat-shock transcription factor HSFA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem Biophys Res Commun 368(3):515–521. https://doi.org/10.1016/j.bbrc.2008.01.134
Chen H, Hwang JE, Lim CJ et al (2010) Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochem Biophys Res Commun 401(2):238–244. https://doi.org/10.1016/j.bbrc.2010.09.038
Guo W, Zhang J, Zhang N et al (2015) The wheat NAC transcription factor TaNAC2L is regulated at the transcriptional and post-translational levels and promotes heat stress tolerance in transgenic Arabidopsis. PLoS ONE 10(8):e0135667. https://doi.org/10.1371/journal.pone.0135667
Gao F, Han X, Wu J et al (2012) A heat-activated calcium-permeable channel–Arabidopsis cyclic nucleotide-gated ion channel 6–is involved in heat shock responses. Plant J 70(6):1056–1069. https://doi.org/10.1111/j.1365-313X.2012.04969.x
Kim JS, Mizoi J, Yoshida T et al (2011) An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol 52(12):2136–2146. https://doi.org/10.1093/pcp/pcr143
Xu ZY, Kim SY, Hyeon do Y et al (2013) The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25(11):4708–4724. https://doi.org/10.1105/tpc.113.119099
Lee S, Seo PJ, Lee HJ et al (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J 70(5):831–844. https://doi.org/10.1111/j.1365-313X.2012.04932.x
Cheong YH, Sung SJ, Kim BG et al (2010) Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in Arabidopsis. Mol Cells 29(2):159–165. https://doi.org/10.1007/s10059-010-0025-z
Li J, Zhou H, Zhang Y et al (2020) The GSK3-like kinase BIN2 is a Molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev Cell 55(3):367–380e6. https://doi.org/10.1016/j.devcel.2020.08.005
Lin H, Yang Y, Quan R et al (2009) Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21(5):1607–1619. https://doi.org/10.1105/tpc.109.066217
Shi H, Lee BH, Wu SJ et al (2003) Overexpression of a plasma membrane Na+/H + antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol 21(1):81–85. https://doi.org/10.1038/nbt766
Jayakannan M, Bose J, Babourina O et al (2013) Salicylic acid improves salinity tolerance in Arabidopsis by restoring membrane potential and preventing salt-induced K + loss via a GORK channel. J Exp Bot 64(8):2255–2268. https://doi.org/10.1093/jxb/ert085
Kim BG, Waadt R, Cheong YH et al (2007) The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J 52(3):473–484. https://doi.org/10.1111/j.1365-313X.2007.03249.x
Yang Z, Wang C, Xue Y et al (2019) Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat Commun 10(1):1199. https://doi.org/10.1038/s41467-019-09181-2
Kugler A, Köhler B, Palme K et al (2009) Salt-dependent regulation of a CNG channel subfamily in Arabidopsis. BMC Plant Biol 9:140. https://doi.org/10.1186/1471-2229-9-140
Singla J, Krattinger SG (2016) Biotic stress resistance genes in wheat. Encyclopedia of Food Grains 388–392. https://doi.org/10.1016/B978-0-12-394437-5.00229-1
Dong J, Chen C, Chen Z (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51(1):21–37. https://doi.org/10.1023/a:1020780022549
Reddy CS, Cho M, Kaul T et al (2023) Pseudomonas fluorescens imparts cadmium stress tolerance in Arabidopsis thaliana via induction of AtPCR2 gene expression. J Genet Eng Biotechnol 21(1):8. https://doi.org/10.1186/s43141-022-00457-7
Zheng Z, Mosher SL, Fan B et al (2007) Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol 7:2. https://doi.org/10.1186/1471-2229-7-2
Lai Z, Vinod K, Zheng Z et al (2008) Roles of Arabidopsis WRKY3 and WRKY4 transcription factors in plant responses to pathogens. BMC Plant Biol 8:68. https://doi.org/10.1186/1471-2229-8-68
Chen C, Chen Z (2002) Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol 129(2):706–716. https://doi.org/10.1104/pp.001057
Cecchini NM, Monteoliva MI, Alvarez ME (2011) Proline dehydrogenase contributes to pathogen defense in Arabidopsis. Plant Physiol 155(4):1947–1959. https://doi.org/10.1104/pp.110.167163
Chin K, DeFalco TA, Moeder W et al (2013) The Arabidopsis cyclic nucleotide-gated ion channels AtCNGC2 and AtCNGC4 work in the same signaling pathway to regulate pathogen defense and floral transition. Plant Physiol 163(2):611–624. https://doi.org/10.1104/pp.113.225680
Noctor G, Foyer CH (1998) ASCORBATE AND GLUTATHIONE: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49:249–279. https://doi.org/10.1146/annurev.arplant.49.1.249
Sharma P, Jha AB, Dubey RS et al (2012) Reactive oxygen species, oxidative damage, and Antioxidative Defense Mechanism in Plants under stressful conditions. J Bot 1–26. https://doi.org/10.1155/2012/217037
Nadarajah KK (2020) ROS Homeostasis in Abiotic stress tolerance in plants. Int J Mol Sci 21(15):5208. https://doi.org/10.3390/ijms21155208
Perez IB, Brown PJ (2014) The role of ROS Signaling in Cross-tolerance from Model to Crop. Front Plant Sci 5:754. https://doi.org/10.3389/fpls.2014.00754
Miller G, Shulaev V, Mittler R (2008) Reactive oxygen signaling and abiotic stress. Physiol Plant 133(3):481–489. https://doi.org/10.1111/j.1399-3054.2008.01090.x
Choudhury FK, Rivero RM, Blumwald E et al (2017) Reactive oxygen species, abiotic stress and stress combination. Plant J 90(5):856–867. https://doi.org/10.1111/tpj.13299
Wang C, Yang A, Yin H et al (2008) Influence of water stress on endogenous hormone contents and cell damage of maize seedlings. J Integr Plant Biol 50(4):427. https://doi.org/10.1111/j.1774-7909.2008.00638.x
Mittler R, Vanderauwera S, Gollery M et al (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9(10):490–498. https://doi.org/10.1016/j.tplants.2004.08.009
Sewelam N, Kazan K, Schenk PM (2016) Global plant stress signaling: reactive oxygen species at the cross-road. Front Plant Sci 7:187. https://doi.org/10.3389/fpls.2016.00187
Hasanuzzaman M, Bhuyan MHMB, Zulfiqar F et al (2020) Reactive oxygen species and antioxidant defense in plants under Abiotic stress: revisiting the crucial role of a Universal Defense Regulator. Antioxid (Basel) 9(8):681. https://doi.org/10.3390/antiox9080681
Chirivì D, Betti C (2023) Molecular Links between Flowering and Abiotic Stress Response: A Focus on Poaceae. Plants (Basel) 12(2):331. https://doi.org10.3390/plants12020331
Shahzad R, Jamil S, Ahmad S et al (2021) Harnessing the potential of plant transcription factors in developing climate resilient crops to improve global food security: current and future perspectives. Saudi J Biol Sci 28(4):2323–2341. https://doi.org/10.1016/j.sjbs.2021.01.028
Yusuf O, Rafii MY, Fatai A et al (2020) Submergence Tolerance in Rice: review of mechanism, breeding and, future prospects. Sustainability 12(4):1632. https://doi.org/10.3390/su12041632
Collins NC, Tardieu F, Tuberosa R (2008) Quantitative trait loci and crop performance under abiotic stress: where do we stand? Plant Physiol 147(2):469–486. https://doi.org/10.1104/pp.108.118117
Feng C, Gao H, Zhou Y et al (2023) Unfolding molecular switches for salt stress resilience in soybean: recent advances and prospects for salt-tolerant smart plant production. Front Plant Sci 14:1162014. https://doi.org/10.3389/fpls.2023.1162014
Sun SK, Xu X, Tang Z et al (2021) A molecular switch in sulfur metabolism to reduce arsenic and enrich selenium in rice grain. Nat Commun 12(1):1392. https://doi.org/10.1038/s41467-021-21282-5
Acknowledgements
This study did not receive any specific grant or financial assistance from funding agencies in the public, commercial, or not-for-profit sectors.
Funding
The authors did not receive support from any organization for the submitted work.
Author information
Authors and Affiliations
Contributions
TD prepared the manuscript draft, tables, and figures; DGD performed the critical review, copy editing, and proofreading; PD conceived and designed the study and performed a literature survey and final review of the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
The authors of this article have no conflicting interests to declare that are relevant to the content. Additionally, this article does not contain any studies involving human participants or animals.
Informed consent
The study does not require human participants and informed consent.
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2023_9051_MOESM1_ESM.docx
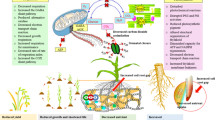
Supplementary Material 1: Fig. 1. Different stress factors are involved in inducing adaptive responses in plants. Fig. 2. Overview of activation of the adaptive mechanism by molecular switches in response to biotic and abiotic stress. Fig. 3. Techniques for the development of stress-resilient crops.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Debnath, T., Dhar, D.G. & Dhar, P. Molecular switches in plant stress adaptation. Mol Biol Rep 51, 20 (2024). https://doi.org/10.1007/s11033-023-09051-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-023-09051-7