Abstract
Key message
The homologous genes OsbHLH068 and AtbHLH112 have partially redundant functions in the regulation of the salt stress response but opposite functions to control flowering in Arabidopsis.
Abstract
The transcription factor (TF) basic/Helix-Loop-Helix (bHLH) is important for plant growth, development, and stress responses. OsbHLH068, which is a homologous gene of AtbHLH112 that is up-regulated under drought and salt stresses, as indicated by previous microarray data analysis. However, the intrinsic function of OsbHLH068 remains unknown. In the present study, we characterized the function and compared the role of OsbHLH068 with that of its homolog, AtbHLH112. Histochemical GUS staining indicated that OsbHLH068 and AtbHLH112 share a similar expression pattern in transgenic Arabidopsis during the juvenile-to-adult phase transition. Heterologous overexpression of OsbHLH068 in Arabidopsis delays seed germination, decreases salt-induced H2O2 accumulation, and promotes root elongation, whereas AtbHLH112 knock-out mutant displays an opposite phenotype. Both OsbHLH068-overexpressing transgenic Arabidopsis seedlings and the Atbhlh112 mutant display a late-flowering phenotype. Moreover, the expression of OsbHLH068-GFP driven by an AtbHLH112 promoter can compensate for the germination deficiency in the Atbhlh112 mutant, but the delayed-flowering phenotype tends to be more severe. Further analysis by microarray and qPCR indicated that the expression of FT is down-regulated in both OsbHLH068-overexpressing Arabidopsis plants and Atbhlh112 mutant plants, whereas SOC1 but not FT is highly expressed in AtbHLH112-overexpressing Arabidopsis plants. A comparative transcriptomic analysis also showed that several stress-responsive genes, such as AtERF15 and AtPUB23, were affected in both OsbHLH068- and AtbHLH112-overexpressing transgenic Arabidopsis plants. Thus, we propose that OsbHLH068 and AtbHLH112 share partially redundant functions in the regulation of abiotic stress responses but have opposite functions to control flowering in Arabidopsis, presumably due to the evolutionary functional divergence of homolog-encoded proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The transcription factor (TF) basic/Helix-Loop-Helix (bHLH) protein comprises a large family in plants (Feller et al. 2011). To date, at least 162 AtbHLHs and 167 OsbHLHs have been identified and can be categorized into 25 subfamilies based on their bHLH domain (Bailey et al. 2003; Heim et al. 2003; Li et al. 2006; Toledo-Ortiz et al. 2003). A typical bHLH domain normally contains two functionally distinct regions: a basic region for DNA binding that recognizes the target cis-acting element, known as E-box (5′-CANNTG-3′), and an HLH region for protein homo- or hetero-dimerization (Feller et al. 2011; Heim et al. 2003; Murre et al. 1989; Pires and Dolan 2010). Notably, a few atypical members known as HLHs lack the basic region. Thus, the HLH–bHLH heterodimeric complex can disrupt the bHLH–bHLH interaction and prevent DNA binding. Based on the dimeric forms, plant bHLH can play a dual role in regulating growth and development. For example, the POSITIVE REGULATOR OF GRAIN LENGTH 1 (PGL1)-ANTAGONIST OF PGL1 (APG) heterodimer, an OsHLH–OsbHLH complex, increases grain length and weight, whereas the APG homodimer decreases grain length and weight (Heang and Sassa 2012). Moreover, plant bHLHs are also involved in many aspects of growth and development, including Z-box binding factor 1 (ZBF1/AtbHLH6), dysfunctional tapetum 1 (DYT1/AtbHLH22), and root hairless 1 (RHL1/OsbHLH3), which are involved in blue light-mediated seedling development, anther development, and root hair development, respectively (Cui et al. 2016; Ding et al. 2009; Maurya et al. 2015). However, the physiological and regulatory roles of most bHLHs in either Arabidopsis or rice remain poorly understood.
Although plants are sessile organisms that cannot escape from deleterious environments, they have already developed and established an exquisite regulatory mechanism to address the changes in their surrounding environment. For example, the inducer of CBF expression 1 (ICE1)-C-repeat binding factor 3/dehydration-responsive element-binding protein 1 A (CBF3/DREB1A) transcriptional cascade, a well-known bHLH–APETALA2/ethylene-responsive factor (AP2/ERF) regulatory pathway, contributes to cold tolerance in Arabidopsis and rice, which illustrates the importance of bHLH in plant stress responses (Chinnusamy et al. 2003; Ito et al. 2006). In fact, AtbHLH116/ICE1 positively regulates the expression of CBF3/DREB1A [AtERF#031 (At4g25480)], which confers freezing tolerance through an ABA-independent pathway (Chinnusamy et al. 2003). In addition to AtbHLH116/ICE1, several plant bHLHs are also involved in abiotic stress responses. For example, OrbHLH1 and OrbHLH2, two ICE-like proteins in wild rice (Oryza rufipogon), confer salt stress tolerance in transgenic Arabidopsis plants through an ICE/CBF-independent and an ABA-independent pathway, respectively (Li et al. 2010; Zhou et al. 2009). OsbHLH062 activates the expression of JA-responsive genes to confer salt stress tolerance, while OsbHLH062 represses the expression of JA-responsive genes when interacting with JASMONATE-ZIM-DOMAIN PROTEIN 9 (OsJAZ9) and NOVEL INTERACTOR OF JAZ (OsNINJA) (Wu et al. 2015). NaCl-induced expression of At bHLH092 confers tolerance to salt and osmotic stresses in Arabidopsis through a pathway that is partially dependent on ABA and SALT OVERLY SENSITIVE 2 (SOS2) (Jiang et al. 2009). In response to the high ambient temperature, AtbHLH9/PHYTOCHROME INTERACTING FACTOR 4 (PIF4) directly activates the expression of YUCCA8 and TAA1, two auxin biosynthetic genes, which in turn triggers hypocotyl elongation in Arabidopsis (for a review, see Proveniers and van Zanten 2013). AtbHLH122 improves drought and osmotic tolerance through direct repression of CYP707A3, an ABA catabolic gene (Liu et al. 2014). Interestingly, AtbHLH112, which belongs to the F subfamily, confers abiotic stress tolerance by increasing proline levels and enhancing reactive oxygen species (ROS) scavenging ability; however, overexpression of AtbHLH112 suppresses lateral root emergence (Liu et al. 2015; Wang et al. 2014). These studies reveal that some bHLHs, such as AtbHLH9/PIF4 and AtbHLH112, are not only involved in the stress response but also play a pleiotropic regulatory role for optimal plant growth and development .
Flowering time is one of the major yield traits correlated with grain production in cereal crops. The precise timing of flowering in plants is coordinately controlled by endogenous and environmental factors, including physiological maturity, accumulated temperature, and day length. In Arabidopsis, a facultative long-day (LD) plant, flowering time is determined by the autonomous, vernalization, photoperiod, and gibberellin pathways (Mouradov et al. 2002), during which photoperiod is the predominant cue controlling flowering time (Andrés and Coupland 2012; Song et al. 2015). These four major pathways integratedly regulate the expression of floral meristem identity genes, such as FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), and LEAFY (LFY), to trigger floral initiation (Wigge et al. 2005). Compared to Arabidopsis, rice is a facultative short-day (SD) plant and thus flowers earlier under SDs than LDs (Hayama et al. 2003). In rice, two FT homologous genes, Heading date 3a (Hd3a) and RICE FLOWERING LOCUS T 1 (RFT1), encode a ‘florigen’ that promotes flowering (Komiya et al. 2008, 2009; Tamaki et al. 2007; Tsuji et al. 2013). The expression of Hd3a and RFT1 is regulated by Early heading date 1 (Ehd1) and Heading date 1 (Hd1) (Doi et al. 2004). In fact, Ehd1 activates the expression of Hd3a and RFT1 under both SDs and LDs, whereas Hd1 functions as a transcriptional activator of Hd3a under SDs but not under LDs (Hayama et al. 2003; Ishikawa et al. 2011). Recently, several plant bHLHs have also been found to be involved in the control of flowering. Four FLOWERING BHLH (FBH1-4) proteins directly activate CONSTANS (CO) expression for photoperiodic flowering in Arabidopsis (Ito et al. 2012). Under SDs, NO FLOWERING IN SHORT DAY (NFL)/bHLH093 promotes flowering through the GA signaling pathway (Sharma et al. 2016). In rice, Oryza sativa late flowering (OsLF), an atypical HLH (OsbHLH119), directly represses Hd1 expression to delay flowering (Wang et al. 2013; Zhao et al. 2011). However, to date, almost no typical OsbHLHs have been shown to regulate flowering.
Flowering time is strongly correlated with environmental factors in plants (for a review, see Riboni et al. 2014). Modification of flowering time for plants to set seeds under a stressful environment is an important survival mechanism for the continuation of the species. Previous studies have indicated that TFs play a vital role in connecting environmental factors to plant flowering. For example, Oryza sativa ABA responsive element binding factor 1 (OsABF1), a drought-inducible bZIP TF, directly activates the expression of OsWRKY104, which suppresses the expression of Edh1 to delay rice flowering under water deficiency (Zhang et al. 2016). Although several OsbHLHs regulate growth, development, or stress responses, the pleiotropic effect of typical OsbHLH on both the stress response and flowering has not yet been reported. In the present study, our data reveal that the function of OsbHLH068 in the regulation of salt stress responses is partially redundant with its homolog, AtbHLH112, but acts oppositely to control flowering in Arabidopsis, presumably due to a functional divergence of the homolog-encoded proteins during evolution.
Results
Salt enhances OsbHLH068 expression
To determine which uncharacterized OsbHLH members may also be involved in regulating abiotic stress responses, we analyzed publicly available microarray data (GSE6901), and several abiotic stress-responsive OsbHLHs with an unknown function were identified (Fig. 1a). Among them, the expression of OsbHLH068 was up-regulated under either drought- or salt-treated conditions. Notably, OsbHLH068 and AtbHLH112 have been categorized as members of the F subfamily (Li et al. 2006). Phylogenetic analysis also revealed that OsbHLH068 was highly homologous to AtbHLH112 based on the sequence similarity within the bHLH domains (Supplementary Fig. S1).
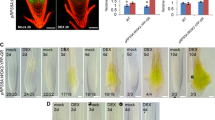
Salt-enhanced OsbHLH068 expression. a Abiotic stress-responsive OsbHLHs. Red and green indicate increased and decreased gene expression, respectively. The scale bar shows log2-fold changes. C, cold treatment; D, drought treatment; H, heat treatment; S, salt treatment. Tos17, rice Tos17 insertion mutant database; TRIM, Taiwan rice insert mutant database. b Histochemical staining of the aerial tissues in OsbHLH068p::GUS transgenic Tainung 67 (Oryza sativa L. spp. japonica cv. Tainung 67) seedlings. c The expression pattern of OsbHLH068 in Nipponbare (Oryza sativa L. spp. japonica cv. Nipponbare) and Tainung 67 aerial tissues by RT-PCR. The Arabic numerals represent the individual rice plants. d Quantification of OsSOS1 and OsbHLH068 mRNA levels in Nipponbare and Tainung 67 aerial tissues by qPCR. The values are the mean ± SD of two independent experiments, each performed in triplicate. *P < 0.05; **P < 0.01, Student’s t-test. Seedlings used for GUS staining (b), RT-PCR (c), and qPCR (d) assays were grown on basal medium for 13 days and then transferred to basal medium containing 0 or 200 mM NaCl for an additional day
To verify whether the expression of OsbHLH068 was in fact up-regulated under salt stress conditions, histochemical staining using OsbHLH068p::GUS transgenic rice was performed. Under 0 mM NaCl conditions, GUS was slightly expressed in the aerial tissues of 14-day-old transgenic seedlings; however, it was enhanced in response to 200 mM NaCl (Fig. 1b). Additionally, the expression pattern of OsbHLH068 was similar to that of OsSOS1, a well-known salt-responsive gene that was up-regulated under 200 mM NaCl conditions in Nipponbare and Tainung 67 seedlings (Fig. 1c, d). These data show that OsbHLH068, an AtbHLH112 homolog, is a salt-responsive gene. Incidentally, we also investigated the spatiotemporal expression of OsbHLH068 in rice. As shown in Fig. 2a, b, the GUS signals were localized to the embryo of germinated transgenic seeds and in the terrestrial tissues of transgenic seedlings. Additionally, GUS signals were also observed in flag leaves, lemma nerves, and anthers during reproductive growth (Fig. 2c).
Overexpression of OsbHLH068 confers salt tolerance in Arabidopsis
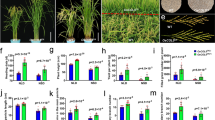
To investigate whether OsbHLH068 plays a positive regulatory role in the salt stress response similar to that of its homolog, AtbHLH112, we compared the seed germination properties of Atbhlh112, Col-0, two OsbHLH068-overexpressing transgenic Arabidopsis lines [35S::OsbHLH068 (6) and (9)], and three independent complemented transformants [AtbHLH112p::GFP-OsbHLH068, AtbHLH112p::OsbHLH068-GFP (10), and (23)] sown on basal medium containing 0 or 100 mM NaCl. Genetic identification of homozygous Atbhlh112 mutant lines (SALK_033618 and _148540), OsbHLH068-overexpressing transgenic Arabidopsis lines, and complemented transformants was performed by genomic DNA genotyping (Supplementary Fig. S2 and S3). Similar to the ABA over-accumulating 35S::LfNCED3 transformant, two independent OsbHLH068-overexpressing transgenic Arabidopsis lines had a lower germination rate in the presence of either 0 or 100 mM NaCl on day 2 compared with the corresponding Col-0 (Fig. 3a). The germination rate of the Atbhlh112 mutant was not significantly different from that of Col-0 in the presence of 0 mM NaCl on day 2 but increased in the mutant compared with Col-0 under 100 mM NaCl conditions (82% and 60%, respectively). On day 3, almost all of the seeds had germinated under 0 mM NaCl conditions; however, two OsbHLH068-overexpressing transgenic lines and the 35S::LfNCED3 transformant still exhibited a lower germination rate under 100 mM NaCl conditions (Supplementary Fig. S4A). Notably, the germination rates of the three independent complemented transformants ranged from approximately 63–72% under 100 mM NaCl conditions on day 2; these rates were similar to the corresponding levels in Col-0 (Fig. 3a). These results indicate that OsbHLH068 can replace the functional role of AtbHLH112 in the regulation of seed germination. On 200 mM NaCl-containing medium, the Atbhlh112 had a higher germination rate than the corresponding Col-0 at days 2 and 3 after seedling, whereas the germination rates of two OsbHLH068-overexpressing transgenic lines were lower than that of the corresponding Col-0 at days 2, 3, 4, and 5 after seedling (Supplementary Fig. S4B). Seed germination reached approximately 100% in each genotype at day 6, reflecting that seed germination delay in OsbHLH068-overexpressing transgenic lines was not due to poor seed vitality. Although Atbhlh112 displayed early germination on 200 mM NaCl-containing medium, more bleached Atbhlh112 seedlings were observed after prolonged culture; however, the late-germinating OsbHLH068-overexpressing transgenic lines had a lower bleached seedling frequency than both Col-0 and Atbhlh112 (Supplementary Fig. S4C, D). Additionally, further investigations revealed that the two OsbHLH068-overexpressing transgenic lines had a lower level of both malondialdehyde (MDA, a lipid peroxidation marker) and hydrogen peroxide (H2O2, a ROS) than the corresponding Col-0 under salt (250 mM NaCl) treatment conditions, whereas the Atbhlh112 mutant had a relatively high level of both MDA and H2O2 (Fig. 3b). Moreover, the levels of both MDA and H2O2 in the complemented transformants were similar to the corresponding levels in Col-0 under salt treatment conditions. Taken together, these data show that OsbHLH068 plays a similar role to that of AtbHLH112 in the regulation of the plant response to salt stress.
Seed germination, physiological investigation, root elongation, and plant morphology in heterologous OsbHLH068-overexpressing transgenic Arabidopsis and the complemented transformant. a The seed germination rates in the Atnced3 mutant, 35S::LfNCED3 transformant, Atbhlh112 mutant, Col-0, two OsbHLH068-overexpressing transgenic Arabidopsis lines, and three complemented transformants on day 2. b The levels of MDA (upper panel) and H2O2 (lower panel) in the Atbhlh112 mutant, Col-0, two OsbHLH068-overexpressing transgenic Arabidopsis lines, and three complemented transformants. c The root lengths of the Atnced3 mutant, 35S::LfNCED3 transformant, Atbhlh112 mutant, Col-0, and two OsbHLH068-overexpressing transgenic Arabidopsis lines. d Phenotypic comparison of the 15-day-old Atnced3 mutant, 35S::LfNCED3 transformant, Atbhlh112 mutant, Col-0, and two OsbHLH068-overexpressing transgenic Arabidopsis lines grown under LD conditions. e The fresh and dry weights of Atnced3 mutant, 35S::LfNCED3 transformant, Atbhlh112 mutant, Col-0, and two OsbHLH068-overexpressing transgenic Arabidopsis lines. *P < 0.05; **P < 0.01, Student’s t-test. The ABA-deficient mutant, Atnced3, and the ABA over-accumulating 35S::LfNCED3 transformant were used as negative and positive controls, respectively. Atbhlh112 mutant, SALK_148540
In addition to the salt stress response, previous studies have shown that overexpression of AtbHLH112 suppresses lateral root emergence and salt-inhibited root growth (Liu et al. 2015; Wang et al. 2014). Therefore, we also investigated whether overexpression of OsbHLH068 affects root development. Indeed, two independent OsbHLH068-overexpressing transgenic Arabidopsis lines and the 35S::LfNCED3 transformant had a longer primary root compared with the corresponding Col-0 seedlings under either 0 or 100 mM NaCl conditions on day 5, whereas the primary roots of both the Atbhlh112 and ABA-deficient Atnced3 mutants were shorter (Fig. 3c). Additionally, the fresh and dry weights of OsbHLH068-overexpressing transgenic Arabidopsis aerial tissues were less than those of Col-0 when grown in soil on day 15 (Fig. 3d, e).
OsbHLH068 and AtbHLH112 act oppositely to control flowering in Arabidopsis
Because OsbHLH068 exerts a pleiotropic effect on many aspects of growth and development in Arabidopsis during the vegetative stage, we subsequently investigated whether OsbHLH068-overexpressing transgenic Arabidopsis had a different phenotype during the reproductive stage. As shown in Fig. 4a, b, the time to flowering in Col-0 was approximately 17 days when grown under LD conditions but was delayed to 21 and 19 days in the 35S::OsbHLH068 (6) and (9) transformants, respectively. Additionally, the flowering times of 35S::OsbHLH068 (6) and the (9) transformants were also delayed when grown under SD conditions (Fig. 4a, b). To validate that the delayed flowering time was not a side effect derived from the constitutive expression of OsbHLH068 in transgenic Arabidopsis seedlings, the spatiotemporal expression of OsbHLH068 was investigated by histochemical staining using OsbHLH068::GUS transgenic Arabidopsis. As shown in Fig. 4c, GUS was expressed in transgenic seedlings during the transition from the vegetative (14 days old) to the reproductive (17 days old) stage. GUS signals were localized only to the cotyledon and its axis in 14-day-old transgenic Arabidopsis seedlings but were also detected in the upper true leaves of 17-day-old transgenic seedlings (Supplementary Fig. S5A). Microscopic observation revealed that GUS signals were localized to the florets, hydathodes, leaf veins, trichome bases, and vascular bundles of the inflorescence in 17-day-old transgenic seedlings (Supplementary Fig. S5B). Because day length plays a key role in determining the flowering time of Arabidopsis, we further examined whether the expression of OsbHLH068 was affected by illumination. As shown in Fig. 4d and Supplementary Fig. S6, the GUS signals in transgenic seedlings decayed when grown in the dark compared with the signals in plants grown under LD conditions. These data suggest that light-induced OsbHLH068 plays a negative role in regulating flowering time in Arabidopsis. Notably, both Atbhlh112 mutants (SALK_033618 and _148540) also displayed a late-flowering phenotype under LD conditions (Fig. 4a, b; Supplementary Fig. S7). These data suggest that OsbHLH068 and AtbHLH112 may act oppositely to control flowering time in Arabidopsis.
Heterologous OsbHLH068-overexpressing transgenic Arabidopsis plants display a late-flowering phenotype. a Phenotypic comparison of the Atbhlh112 mutant, Col-0, and two 35S::OsbHLH068 transformants grown under LD (left) and SD (right) conditions. Seedlings were grown under LD and SD conditions for 19 and 41 days, respectively. b The bolting time of the Atbhlh112 mutant, Col-0, and two 35S::OsbHLH068 transformants grown under LD (left) and SD (right) conditions. The values are the mean ± SD of two independent experiments, each with ten replicates. *P < 0.05; **P < 0.01, Student’s t-test. c Histochemical staining of OsbHLH068p::GUS transgenic Arabidopsis aerial tissues during the vegetative, transitional, and reproductive stages. Seedlings were grown under LD conditions. Scale bar, 1 cm. Enlarged versions of each black-disc-labeled image are shown in Supplementary Fig. S5a. d Histochemical staining of dark- and LD-treated OsbHLH068p::GUS transgenic aerial tissues over time. Seedlings were grown under LD conditions for 11 days and then transferred to darkness or constant LD conditions for an additional 3 days. Scale bar, 1 cm. Enlarged versions of each black disc-labeled image are shown in Supplementary Fig. S6. Atbhlh112 mutant, SALK_148540
To confirm that OsbHLH068 and AtbHLH112 act oppositely to control flowering time, we subsequently investigated the flowering times of Atbhlh112, Col-0, and three complemented transformants under LD conditions. Indeed, the flowering times of the three complemented transformants were more severely delayed than Atbhlh112 when compared to Col-0 seedlings under LD conditions (Fig. 5a, b). Additionally, GUS driven by a 2.2-kb AtbHLH112 native promoter, as used in the complemented transformants expressing GFP-OsbHLH068 or OsbHLH068-GFP, was expressed in the AtbHLH112::GUS transformant during the transition from the vegetative (14 days old) to the reproductive (17 days old) stage (Fig. 5c). Similar to OsbHLH068::GUS transgenic Arabidopsis plants, the GUS signals were localized to the cotyledon and its axis in 14-day-old AtbHLH112::GUS seedlings and presented in the upper true leaves of 17-day-old AtbHLH112::GUS seedlings (Supplementary Fig. S5A vs. S8A). Microscopic observation revealed GUS signal localization in the florets, leaf veins, and trichome bases of 17-day-old AtbHLH112::GUS seedlings (Supplementary Fig. S8B). Incidentally, both the GFP-OsbHLH068 and OsbHLH068-GFP fusion proteins were localized to the nucleus of root cells in the complemented transformants (Fig. 5d). Taken together, these data indicate that OsbHLH068 and AtbHLH112 share a similar expression pattern during the phase transition from vegetative to reproductive growth but act oppositely to control the flowering time.
Complementary expression of GFP-OsbHLH068 or OsbHLH068-GFP causes a more severe late-flowering phenotype in the Atbhlh112 mutant background grown under LD conditions. a Phenotypic comparison of Col-0, the Atbhlh112 mutant, and the three complemented transformants. The seedlings were grown in soil for 19 days. b The bolting time of Col-0, the Atbhlh112 mutant, and the three complemented transformants. The values are the mean ± SD of two independent experiments, each with ten replicates. *P < 0.05; **P < 0.01, Student’s t-test. c Histochemical staining of the aerial tissues in AtbHLH112p::GUS transgenic seedlings during vegetative, transitional, and reproductive stages. Seedlings were grown in soil for the indicated period of time. Scale bar 1 cm. Enlarged versions of each black disc-labeled image are shown in Supplementary Fig. S8A. d Subcellular localization of GFP-OsbHLH068 (left panel) and OsbHLH068-GFP (right panel) proteins in complemented transformants. Atbhlh112 mutant, SALK_148540
Transcriptomic analysis of OsbHLH068-overexpressing transgenic Arabidopsis plants
To better understand the molecular mechanism of OsbHLH068 in the regulation of the salt stress response and flowering, we conducted a comparative transcriptomic analysis of OsbHLH068-overexpressing transformant [35S::OsbHLH068 (9)] and Col-0 seedlings. After background correction and normalization, a total of 568 differentially expressed probes (DEPs) were identified. Among these, 168 DEPs were up-regulated while 400 DEPs were down-regulated in OsbHLH068-overexpressing transformants (Fig. 6a; Supplementary Data 1). As expected, several well-known stress-responsive genes were found among these DEPs, including MYB domain protein 4 (MYB4), protein phosphatase 2 C 5 (PP2C5), CBF2, and 1-aminocyclopropane-1-carboxylate synthase 11 (ACS11). Notably, almost none of the flowering-related genes, such as LFY, SOC1, and FT, were detected as DEPs. However, the log2-fold changes in SOC1 and FT were approximately −0.25 and −0.79, respectively, with a P-value of at least < 0.05 based on Student’s t-test (Fig. 6c, Supplementary Data 1). Thus, a qPCR assay was performed to verify the microarray data. As shown in Fig. 6b, d, the mRNA levels of MYB4 and PP2C5 were higher in OsbHLH068-overexpressing transformants than in Col-0, while the mRNA levels of CBF2, ACS11, SOC1, and FT were lower in OsbHLH068-overexpressing transformants than in Col-0. The results obtained by qPCR were consistent with the microarray data. In fact, the qPCR data also revealed that the late-flowering Atbhlh112 mutant had a relatively low level of both FT and APETALA1 (AP1) expression and a relatively high level of FLOWERING LOCUS C (FLC) expression, which were similar to those seen in the OsbHLH068-overexpressing transformants (Fig. 6d; Supplementary Fig. S9).
Transcriptomic analysis of the OsbHLH068-overexpressing transformant and Col-0. a Identification of DEPs. Red and green spots indicate up- and down-regulated DEPs, respectively. OsbHLH068-related DEPs are documented in Supplementary Data 1. b Quantification of MYB4, PP2C5, CBF2, and ACS11 mRNAs in two OsbHLH068-overexpressing transgenic Arabidopsis lines and Col-0 by qPCR. c Microarray expression analysis of LFY, SOC1, and FT in OsbHLH068-overexpressing transgenic Arabidopsis plants. The scale bar shows log2-fold changes. Red and green colors indicate increased and decreased gene expression. d Quantification of LFY, SOC1, and FT mRNAs in Atbhlh112, Col-0, and two OsbHLH068-overexpressing transgenic Arabidopsis lines by qPCR. The total RNA used in the microarray and qPCR assays was extracted from the same aerial tissues of 17-day-old seedlings. The values presented in b and d are the mean ± SE of 4 biological replicates, each with two technical replicates. *P < 0.05; **P < 0.01, Student’s t-test. Atbhlh112 mutant, SALK_148540
Common differentially expressed genes between OsbHLH068- and AtbHLH112-overexpressing Arabidopsis
Because OsbHLH068 and AtbHLH112 share a highly similar bHLH domain, we assumed that both TFs should commonly regulate the expression of certain downstream genes, especially when they were constitutively expressed in the same genetic background. Therefore, we further comparatively analyzed the publicly available microarray data, GSE84087 and GSE60260, to identify the common targets between OsbHLH068- and AtbHLH112-overexpressing transgenic Arabidopsis. After background correction and normalization of AtbHLH112-related microarray data (GSE60260), a total of 11,616 DEPs consisting of 6632 up-regulated and 4984 down-regulated DEPs were identified (Supplementary Data 2). Notably, a total of 358 DEPs were commonly presented in both OsbHLH068- and AtbHLH112-overexpressing transgenic Arabidopsis (Fig. 7), of which 61 and 263 DEPs were identically up- and down-regulated, respectively, in both transgenic plants. Additionally, the 61 common up-regulated DEPs can be reflected in 52 genes (denoted as common up-regulated DEGs) based on TIGR annotation, while the 263 common down-regulated DEPs can be represented in 206 genes (designated as common down-regulated DEGs), including CBF2 and ACS11 (Supplementary Data 3). Gene ontology analysis revealed that the major ontological categories of the commonly regulated DEGs were ‘response to stimulus’ and ‘transcription regulator activity’ of ‘biological process’ and ‘molecular function’, respectively (Supplementary Fig. S10). Incidentally, further promoter analysis indicated that 51 and 194 common up- and down-regulated DEGs, respectively, contain at least one E-box element in their 1-kb promoter region (Supplementary Data 4).
Discussion
Partial functional redundancy of AtbHLH112 and OsbHLH068 in the stress response
The phytohormone abscisic acid (ABA) plays an important role in regulating seed maturation, dormancy, stomatal closure, and abiotic stress responses (Gubler et al. 2005; Karssen et al. 1983; Leung and Giraudat 1998; MacRobbie 1998; Seiler et al. 2011). In fact, ABA-mediated seed dormancy is an adaptive mechanism that maintains viable seeds in a quiescent state and leads to the escape from or avoidance of a stressful environment (Seo et al. 2006). Indeed, overexpression of ABA biosynthetic genes, such as nine-cis-epoxycarotenoid dioxygenase 3 (AtNCED3) and ABA2, delays seed germination and confers drought/salt stress tolerance in transgenic Arabidopsis plants (Cheng et al. 2002; Iuchi et al. 2001; Lin et al. 2007). In contrast, ABA-deficient mutants, such as aba1, aba2, and aba4, have a rapid seed germination phenotype but are susceptible to osmotic stress (Lin et al. 2007; North et al. 2007). Additionally, root architecture alteration is another adaptive mechanism for plants to survive under stress conditions (Price et al. 1997; Serraj et al. 2004; Uga et al. 2013). In rice, drought-resistant varieties usually develop a deeper root system so that the roots can take up water from deeper soil layers to manage water deficiency (Gowda et al. 2011; Price et al. 1997; Uga et al. 2013). As mentioned in a previous study, AtbHLH112 confers abiotic stress tolerance by enhancing the ROS scavenging ability and promotes primary root growth under salt-treated conditions (Liu et al. 2015). In this study, heterologous overexpression of OsbHLH068, an AtbHLH112 homolog, in Arabidopsis delayed seed germination, decreased the accumulation of MDA and H2O2, and enhanced primary root elongation under salt-treated conditions, whereas the Atbhlh112 mutant displayed a rapid seed germination phenotype, a relatively high level of MDA and H2O2, and a short root length phenotype (Fig. 3a–c and Supplementary Fig. S4). Notably, the seed germination and root elongation properties of OsbHLH068-overexpressing transgenic Arabidopsis plants and the Atbhlh112 mutant were highly similar to those of the ABA over-accumulating 35S::LfNCED3 transformant and the ABA-deficient mutant, Atnced3, respectively. Furthermore, complementary expression of either GFP-OsbHLH068 or OsbHLH068-GFP driven by a 2.2-kb AtbHLH112 promoter could restore, partially or completely, the early germination and the H2O2 over-accumulation of the Atbhlh112 mutant to normal germination and accumulation, respectively (Fig. 3a, b). These data showed that the regulatory role of AtbHLH112 in seed germination and H2O2 scavenging could be replaced by OsbHLH068, presumably due to the conserved function between Arabidopsis (dicot) and rice (monocot). Additionally, comparative transcriptomic analysis indicated that 52 up-regulated and 206 down-regulated DEGs were commonly presented in OsbHLH068- and AtbHLH112-overexpressing transgenic Arabidopsis plants (Supplementary Data 3), of which 51 common up-regulated DEGs and 194 common down-regulated DEGs had at least one E-box element (5′-CANNTG-3′) in their 1-kb promoter region, including several well-known stress-responsive genes [e.g., AtERF15 (At2g31230), AtPUB23 (At2g35930) and WRKY48 (At5g49520)] (Supplementary Data 4). AtERF15, a commonly up-regulated DEG, plays a positive role in regulating ABA-mediated drought tolerance, whereas plant U-box 23 (AtPUB23), a commonly down-regulated DEG, negatively regulates ABA-mediated drought stress responses (Lee et al. 2015; Seo et al. 2012). The osmotic stress- and pathogen-induced AtWRKY48, a common down-regulated DEG, functions as a negative regulator of defense-related genes, such as PRs, and basal resistance to the bacterial pathogen Pseudomonas syringae (Xing et al. 2008). Taken together, our data reveal that OsbHLH068 plays a role similar to that of AtbHLH112 in the regulation of abiotic stress responses, presumably due to the partially functional redundancy of these homologous genes.
Functional divergence of AtbHLH112 and OsbHLH068 in flowering control
Homologous gene-encoded proteins usually play a similar role in regulating plant developmental and physiological processes. For example, the CBFs/DREBs, which belong to the AP2/ERF family, play a positive regulatory role in the cold tolerance of evolutionarily diverse plant species, including Arabidopsis and rice (for a review, see Chinnusamy et al. 2010). Notably, both OsbHLH068 and AtbHLH112 are members of the F subfamily (Li et al. 2006). In the F subfamily, both the OsbHLH068 domain and the AtbHLH112 domain share a high sequence similarity (Supplementary Fig. S1). Additionally, OsbHLH068 and AtbHLH112 are expressed in the upper true leaves of transgenic Arabidopsis seedlings in a nearly overlapping pattern during the juvenile-to-adult phase (Fig. 4c vs. 5c, Supplementary Fig. S5 vs. S8). These biological characteristics suggest that OsbHLH068 and AtbHLH112 should play similar roles in regulating plant growth and development, especially during the phase transition process. Indeed, heterologous overexpression of OsbHLH068 in Arabidopsis delayed flowering with a relatively low level of SOC1, FT, or AP1 expression and a relatively high level of FLC expression under LD conditions (Figs. 4a, b, 6c, d; Supplementary Fig. S9). Surprisingly, two Atbhlh112 mutant lines (SALK_033618 and _148540) also displayed a late-flowering phenotype under LD conditions (Fig. 4a, b; Supplementary Fig. S7). Interestingly, the late-flowering Atbhlh112 mutant (SALK_148540) exhibited a relatively low level of both FT and AP1 expression and a relatively high level of FLC expression under LD conditions, whereas constitutive expression of AtbHLH112 in Arabidopsis up-regulated the expression of SOC1 but not FT (Fig. 6d; Supplementary Fig. S9 and Data 2). Inconceivably, complementary expression of GFP-OsbHLH068 or OSbHLH068-GFP driven by a 2.2-kb AtbHLH112 promoter in the late-flowering Atbhlh112 mutant delayed the flowering time more severely under LD conditions (Fig. 5a, b). The opposite effect of OsbHLH068 and AtbHLH112 on flowering control did not appear to be caused by the differential transcriptional activity because complementary expression of GFP-OsbHLH068 or OsbHLH068-GFP driven by a 2.2-kb AtbHLH112 promoter in the Atbhlh112 mutant partially or completely restored the early seed germination and H2O2 over-accumulation to normal levels (Fig. 3a, c). Therefore, we assume that OsbHLH068 and AtbHLH112 act oppositely in flowering control, presumably due to the divergent evolution of plant flowering.
Divergent evolution is a pathway involved in speciation that contributes to the species abundance and diversity of biological systems. Gene duplication, followed by the functional divergence of paralog-encoded proteins, is considered to be a major driving force in the evolution of biological diversity (for review, see Kondrashov et al. 2002; Ohno 1970). For example, ATX1 and ATX2, two Arabidopsis thaliana TRITHORAX homologous genes derived from a segmental duplication, belong to the same clade as the sister paralogs (Alvarez-Venegas and Avramova 2002; Baumbusch et al. 2001). Although the structure of ATX1 is similar to that of ATX2, ATX1 and ATX2 have opposite biochemical activities (Saleh et al. 2008): ATX1 trimethylates K4 of histone H3, whereas ATX2 dimethylates it. They activate and inactivate the transcriptional expression of common target genes, including WRKY70, a TF gene involved in the regulation of the disease response. In addition to ATX1 and ATX2, the ARABIDOPSIS MYOTUBULARIN1 (AtMTM1) and AtMTM2 homologs also originated from a segmental duplication and encode catalytically active enzymes with a similar domain architecture and a conserved biochemically active catalytic site (Ding et al. 2012). However, AtMTM1 elevates the cellular level of phosphatidylinositol 5-phosphate (PtdIns5P) in response to dehydration stress, but the function of AtMTM2 remains unclear. More importantly, AtMTM1-mediated PtdIns5P can bind to the PHD domain of ATX1 and then repress the methylation activity of ATX1 (Alvarez-Venegas et al. 2006a, b; Ndamukong et al. 2010). These cases illustrate that the functional divergence of paralog-encoded proteins confers, at least in part, the biochemical and biological diversity of plants. To identify flowering-related genes in monocot rice, a facultative SD plant, the transcriptional expression of two FT homologous genes, Hd3a and RFT1 paralogs, is activated by Ehd1 in the vascular tissues of leaf blades under both SDs and LDs (Doi et al. 2004). However, the transcriptional expression of Hd3a is repressed by Hd1, a CO homolog, under LDs (Kojima et al. 2002). Notably, no Ehd1 homolog was found in Arabidopsis. In contrast, the dicot Arabidopsis is classified as a facultative LD plant. Transcriptional expression of FT is activated by CO in the vascular tissues of leaf blades under LDs (Kardailsky et al. 1999). Interestingly, the transcriptional regulation of homologous Hd3a and FT by Hd1 and CO, two homologous zinc-finger TFs, also displays an opposite relationship in the flowering control between rice and Arabidopsis, respectively, under LDs. These data indicate that functional divergence of homologous flowering-related genes was present during the divergent evolution of rice and Arabidopsis. The present data indicate that OsbHLH068 and AtbHLH112 have a similar transcriptional pattern during the juvenile-to-adult phase transition, but both OsbHLH068-overexpressing transgenic Arabidopsis plants and the Atbhlh112 mutants show a late-flowering phenotype with a relatively low level of FT expression under LDs. Notably, constitutive expression of AtbHLH112 in transgenic Arabidopsis seedlings resulted in a relatively high expression level of SOC1 but no difference from that of FT (Supplementary Data 2). Further promoter analysis revealed that at least 5, 3, 4, and 6 E-box elements were located in the promoter region within 1 kb upstream of the FT, SOC1, Hd3a, and RFT transcription start sites, respectively (Supplementary Table 1). Thus, we conclude that OsbHLH068 and AtbHLH112, two homologous TFs, have an opposite effect on the transcriptional regulation of downstream homologous flowering genes between rice and Arabidopsis, which is similar to the effect observed for Hd1 and CO in the transcriptional regulation of Hd3a and FT, respectively. Additionally, AtbHLH112 seems to play a fine-tuning role in regulating flowering time by interacting with the different proteins in Arabidopsis. Taken together, we propose that the opposite roles of OsbHLH068 and AtbHLH112 in flowering control, as well as Hd1 and CO, are to be at least partially incorporated into the divergent evolution of rice and Arabidopsis (for a summary, see Fig. 8).
A putative role of OsbHLH068 in salt stress response and flowering
Flowering is important for plants to complete the life cycle. However, the timing of flowering is highly susceptible to environmental conditions, particularly when the plant is exposed to abiotic stresses. In fact, many studies have shown that the effects of abiotic stress on flowering control are different in various plant species. For example, the flowering of Arabidopsis plants is accelerated by drought and delayed by salinity (Kazan and Lyons 2016; Riboni et al. 2014), whereas the flowering of rice plants is delayed by drought (Galbiati et al. 2016). Notably, previous studies have also shown that certain stress-inducible TFs play important roles in connecting environmental factors to flowering, including CmMYB2 and OsABF1. Heterologous overexpression of CmMYB2, an abiotic stress-inducible R2R3-MYB TF in chrysanthemum, improves osmotic-stress tolerance and delays flowering time in transgenic Arabidopsis plants (Shan et al. 2012). In rice, a drought-inducible bZIP TF, OsABF1, acts as a negative regulator of floral transition upon water deficit (Zhang et al. 2016). In this study, our data reveal that heterologous overexpression of OsbHLH068, a salt-inducible TF, in Arabidopsis results in decreased salt-dependent accumulation of either MDA or H2O2, increased root length, and delayed flowering. In fact, the OsbHLH068-overexpressing transgenic rice plants also displayed a late-flowering phenotype with a relatively low level of Hd3a and RFT1 expression (unpublished data). Thus, we speculate that OsbHLH068 may play a pivotal role in linking the salt stress response to rice flowering.
Materials and methods
Plant materials and growth conditions
In Arabidopsis, the SALK_033618 and _148540 lines are AtbHLH112 knock-out mutants, which have been documented in a previous study (Wang et al. 2014). These mutants were obtained from the Arabidopsis Biological Resource Center (ABRC, OH). All mutants were derived from the Columbia-0 (Col-0) accession. Seeds were treated with 1.2% (v/v) commercial bleach for 15 min, rinsed twice with sterile water for 10 min each, and subsequently stored at 4 °C in the dark for 3 days. Sterilized and cold-pretreated seeds were sown on agar plates or in pots containing disinfected Tref substrate (Jiffy). Seedlings were grown at 24 °C under LD (16-h light/8-h dark cycle) or SD (8-h light/16-h dark cycle) conditions with a light intensity of approximately 100 µE/s m2. For the germination and root elongation tests, the basal agar medium was composed of half-strength MS salts (Murashige and Skoog 1962), B5 organic compounds (Gamborg et al. 1968), 0.05% MES [2-(N-morpholino)ethane sulfonic acid monohydrate], and 1% sucrose. The Atnced3 mutant and 35S::LfNCED3 transformant were obtained from Dr. Wan-Hsing Cheng (Institute of Plant and Microbial Biology, Academia Sinica, Taipei, Taiwan) and have been used in previous studies (Chen et al. 2011; Wan and Li 2006).
In rice, Oryza sativa L. cv. Nipponbare and Oryza sativa L. cv. Tainung 67 were investigated in the present analysis. The seeds were sterilized and imbibed at 37 °C for 2 days in the dark. After imbibition, the germinated seeds were grown on a wired stand in beakers at 28 °C under LD conditions (16-h light/8-h dark cycle) with a light intensity of approximately 100 µE/s m2. The basal medium was Kimura B solution (Yoshida et al. 1976).
Transgene constructs
The full-length coding sequences of OsbHLH068 and GFP, with or without a stop codon, were PCR-amplified and cloned into the pGEM-T Easy vector (Promega). These fragments were then subcloned into a binary vector, pCAMBIA-1300, where their expression was driven by either a 2.2-kb AtbHLH112 native promoter (AtbHLH112p::GFP-OsbHLH068 and AtbHLH112p::OsbHLH068-GFP) or a CaMV 35 S promoter (35S::OsbHLH068). After sequencing, the AtbHLH112p::GFP-OsbHLH068 and AtbHLH112p::OsbHLH068-GFP constructs were subsequently transformed into the Atbhlh112 (SALK_148540) mutant by the floral dipping method for functional complementation and protein subcellular localization assays, and the 35S::OsbHLH068 construct was introduced into Columbia-0 for functional analysis. Additionally, both 1.2-kb OsbHLH068 and 2.2-kb AtbHLH112 native promoters were used to replace the 35 S promoter to drive the expression of the β-glucuronidase (GUS) reporter gene in the binary vector pCAMBIA-1305.1. After sequencing, the OsbHLH068p::GUS and AtbHLH112p::GUS constructs were subsequently transformed into Col-0 and/or Oryza sativa L. cv. Tainung 67 via an Agrobacterium-mediated method for analyzing spatiotemporal gene expression.
Phenotypic comparisons and plant weight measurements
For the phenotypic comparisons, cold-pretreated seeds from each type of Arabidopsis plant were grown in pots containing disinfected Tref substrate (Jiffy). For the plant weight measurements, seedlings were grown under LD conditions. The aerial parts of the 15-day-old plants were excised and the detached rosette leaves were used to measure the fresh weight. These detached tissues were subsequently vacuum dried at least overnight to measure the dry weight. All fresh and dry weights are the mean ± SD of two independent experiments, each with 6 biological replicates (each with an average value of two technical replicates).
Germination and root elongation tests
For the germination and root elongation tests, cold-pretreated seeds of each type of Arabidopsis plant were sown on agar plates containing basal agar medium with or without 100 mM NaCl. Germination was defined as the point when radicle emergence first started to exceed seed coat. All germination rates are the mean ± SE of 4 independent biological replicates. Each biological replicate contains at least 35 seeds that were harvested from an individual Arabidopsis plant. The root length was measured after vertical growth for 5 days on agar plates. All root lengths are the mean ± SE of 20 independent biological replicates.
MDA and H2O2 determinations
For the MDA and H2O2 assays, cold-pretreated seeds of each type of Arabidopsis plant were grown in pots containing disinfected Tref substrate (Jiffy) under LD conditions. After growing for 12 days in pots, the seedlings were watered with fresh water containing 0 or 250 mM NaCl for an additional 3 days and then harvested for analysis. The MDA and H2O2 levels were measured according to methods described by Heath and Packer (1968), Jana and Choudhuri (1981), respectively.
RNA extraction and cDNA synthesis
Total RNA was extracted from the various plant tissues using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. To avoid genomic DNA contamination, total RNA was treated with Turbo DNA-free™ DNase (Ambion) following the manufacturer’s instructions. The DNase-treated total RNA was subjected to cDNA synthesis using the SuperScript™ III first-strand synthesis system (Invitrogen) according to the manufacturer’s instructions.
Reverse transcription-PCR (RT-PCR) and quantitative PCR (qPCR)
The initial amount of template cDNA in each RT-PCR and qPCR reaction was 25 and 10 ng, respectively. The qPCR reactions were performed using an ABI 7500 system with the SYBR® Green PCR Master Mix Kit [Applied Biosystems (ABI)]. The 2−ΔCT was used to show a difference between the target gene and internal control. OsACTIN1 was used as an internal control for qPCR normalization. The primer sequences used are listed in Supplementary Table 2.
Spatiotemporal gene expression and protein subcellular localization
Samples were soaked in fixation buffer (0.3% formaldehyde, 10 mM MES hydrate, 0.3 M mannitol, and 2 mM dithiothreitol, pH 5.6) for 30 min before staining. After fixation, the samples were rinsed twice with 50 mM sodium phosphate (pH 7.0) and then submerged in a staining solution [50 mM sodium phosphate dibasic, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 2 mM dithiothreitol, and 1 mM X-Gluc (5-bromo-4-chloro-3-indolyl β-D-glucuronide cyclohexylammonium salt), pH 7.0]. The staining assay was conducted under ambient conditions. For the protein subcellular localization assay, GFP fluorescence was detected with spectral settings of 500–540 nm for emission and 488 nm for excitation using a Zeiss LSM 510 Meta confocal microscope.
Microarray and data analysis
Total RNA was amplified and labeled using a Low-Input Quick Amp Labeling Kit, One-Color (Agilent, USA), according to the manufacturer’s instructions. Cyanine 3 (Cy3)-labeled cRNA was fragmented by incubation at 60 °C for 30 min. After fragmentation, Cy3-labeled cRNA was pooled and hybridized to the Agilent Arabidopsis V4 Oligo 4 × 44 K Microarray as suggested by the manufacturer. The array image was analyzed using Feature Extraction software version 10.7.1.1 with default settings. The gene expression data are available under accession number GSE84087. Additionally, the microarray dataset (GSE60260) was obtained from the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/). To identify DEPs, the raw microarray data were analyzed with the Bioconductor Limma package (Ritchie et al. 2015). The raw data were background-corrected using the ‘normexp’ method and then normalized using the ‘quantile’ method. Each up- or down-regulated DEP had a log2-fold change > or <1, respectively, with a P-value < 0. 05 based on Student’s t-test. Each DEP-annotated gene is listed and described in the corresponding supplemental data. Common DEG ontology graphical analysis was conducted using the agriGO database (Du et al. 2010).
References
Alvarez-Venegas R, Avramova Z (2002) SET-domain proteins of the Su(var)3–9, E(z) and Trithorax families. Gene 285:25–37. doi:10.1016/s0378-1119(02)00401-8
Alvarez-Venegas R, Sadder M, Hlavacka A, Baluska F, Xia Y, Lu G, Firsov A, Sarath G, Moriyama H, Dubrovsky JG, Avramova Z (2006a) The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc Natl Acad Sci U S A 103:6049–6054. doi:10.1073/pnas.0600944103
Alvarez-Venegas R, Xia Y, Lu G, Avramova Z (2006b) Phosphoinositide 5-phosphate and phosphoinositide 4-phosphate trigger distinct specific responses of Arabidopsis genes. Plant Signal Behav 1:140–151. doi:10.4161/psb.1.3.2997
Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13:627–639. doi:10.1038/nrg3291
Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, Jakoby M, Werber M, Weisshaar B (2003) Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15:2497–2502. doi:10.1105/tpc.151140
Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB (2001) The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29:4319–4333. doi:10.1093/nar/29.21.4319
Chen HC, Hwang SG, Chen SM, Shii CT, Cheng WH (2011) ABA-mediated heterophylly is regulated by differential expression of 9-cis-epoxycarotenoid dioxygenase 3 in lilies. Plant Cell Physiol 52:1806–1821. doi:10.1093/pcp/pcr117
Cheng W-H, Endo A, Zhou L, Penney J, Chen H-C, Arroyo A, Leon P, Nambara E, Asami T, Seo M (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14:2723–2743
Chinnusamy V, Ohta M, Kanrar S, Lee B-H, Hong X, Agarwal M, Zhu J-K (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043–1054
Chinnusamy V, Zhu J-K, Sunkar R (2010) Gene regulation during cold stress acclimation in plants. Methods in Mol Biol 639:39–55. doi:10.1007/978-1-60761-702-0_3
Cui J, You C, Zhu E, Huang Q, Ma H, Chang F (2016) Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. Plant Cell 28:1078–1093. doi:10.1105/tpc.15.00986
Ding W, Yu Z, Tong Y, Huang W, Chen H, Wu P (2009) A transcription factor with a bHLH domain regulates root hair development in rice. Cell Res 19:1309–1311. doi:10.1038/cr.2009.109
Ding Y, Ndamukong I, Zhao Y, Xia Y, Riethoven J-J, Jones DR, Divecha N, Avramova Z (2012) Divergent functions of the myotubularin (MTM) homologs AtMTM1 and AtMTM2 in Arabidopsis thaliana: evolution of the plant MTM family. Plant J 70:866–878. doi:10.1111/j.1365-313x.2012.04936.x
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A (2004) Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev 18:926–936. doi:10.1101/gad.1189604
Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38:W64–W70. doi:10.1093/nar/gkq310
Feller A, Machemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66:94–116. doi:10.1111/j.1365-313x.2010.04459.x
Galbiati F, Chiozzotto R, Locatelli F, Spada A, Genga A, Fornara F (2016) Hd3a, RFT1 and Ehd1 integrate photoperiodic and drought stress signals to delay the floral transition in rice. Plant Cell Environ 39:1982–1993. doi:10.1111/pce.12760
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151–158. doi:10.1016/0014-4827(68)90403-5
Gowda VRP, Henry A, Yamauchi A, Shashidhar HE, Serraj R (2011) Root biology and genetic improvement for drought avoidance in rice. Field Crops Res 122:1–13. doi:10.1016/j.fcr.2011.03.001
Gubler F, Millar AA, Jacobsen JV (2005) Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8:183–187. doi:10.1016/j.pbi.2005.01.011
Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K (2003) Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422:719–722. doi:10.1038/nature01549
Heang D, Sassa H (2012) Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS One 7:e31325. doi:10.1371/journal.pone.0031325
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125:189–198. doi:10.1016/0003-9861(68)90654-1
Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20:735–747. doi:10.1093/molbev/msg088
Ishikawa R, Aoki M, Kurotani K-I, Yokoi S, Shinomura T, Takano M, Shimamoto K (2011) Phytochrome B regulates heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol Genet Genomics 285:461–470. doi:10.1007/s00438-011-0621-4
Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47:141–153. doi:10.1093/pcp/pci230
Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, Imaizumi T (2012) FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc Natl Acad Sci 109:3582–3587. doi:10.1073/pnas.1118876109
Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27:325–333. doi:10.1046/j.1365-313x.2001.01096.x
Jana S, Choudhuri MA (1981) Glycolate metabolism of three submersed aquatic angiosperms during ageing. Aquat Bot 12:345–354. doi:10.1016/0304-3770(81)90047-4
Jiang Y, Yang B, Deyholos MK (2009) Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol Genet Genomics 282:503–516. doi:10.1007/s00438-009-0481-3
Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286:1962–1965. doi:10.1126/science.286.5446.1962
Karssen CM, van der Swan DLC, Breekland AE, Koornneef M (1983) Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157:158–165. doi:10.1007/bf00393650
Kazan K, Lyons R (2016) The link between flowering time and stress tolerance. J Exp Bot 67:47–60. doi:10.1093/jxb/erv441
Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M (2002) Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol 43:1096–1105. doi:10.1093/pcp/pcf156
Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K (2008) Hd3a and RFT1 are essential for flowering in rice. Development 135:767–774. doi:10.1242/dev.008631
Komiya R, Yokoi S, Shimamoto K (2009) A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136:3443–3450. doi:10.1242/dev.040170
Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV (2002) Selection in the evolution of gene duplications. Genome Biol. doi:10.1186/gb-2002-3-2-research0008
Lee S-B, Lee S-J, Kim SY (2015) AtERF15 is a positive regulator of ABA response. Plant Cell Rep 34:71–81. doi:10.1007/s00299-014-1688-2
Leung J, Giraudat J (1998) Abscisic acid signal transduction. Ann Rev Plant Physiol Plant Mol Biol 49:199–222. doi:10.1146/annurev.arplant.49.1.199
Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D (2006) Genome-wide analysis of basic/Helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol 141:1167–1184. doi:10.1104/pp.106.080580
Li F, Guo S, Zhao Y, Chen D, Chong K, Xu Y (2010) Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from Dongxiang wild rice confers freezing and salt tolerance in transgenic Arabidopsis. Plant Cell Rep 29:977–986. doi:10.1007/s00299-010-0883-z
Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, Cheng WH (2007) Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol 143:745–758. doi:10.1104/pp.106.084103
Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, Li W-X (2014) bHLH122is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol 201:1192–1204. doi:10.1111/nph.12607
Liu Y, Ji X, Nie X, Qu M, Zheng L, Tan Z, Zhao H, Huo L, Liu S, Zhang B, Wang Y (2015) Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol 207:692–709. doi:10.1111/nph.13387
MacRobbie EAC (1998) Signal transduction and ion channels in guard cells. Philosoph Trans Roy Soc B Biol Sci 353:1475–1488. doi:10.1098/rstb.1998.0303
Maurya JP, Sethi V, Gangappa SN, Gupta N, Chattopadhyay S (2015) Interaction of MYC2 and GBF1 results in functional antagonism in blue light-mediated Arabidopsis seedling development. Plant J 83:439–450. doi:10.1111/tpj.12899
Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14:S111–S130
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497. doi:10.1111/j.1399-3054.1962.tb08052.x
Murre C, McCaw PS, Baltimore D (1989) A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777–783. doi:10.1016/0092-8674(89)90682-x
Ndamukong I, Jones DR, Lapko H, Divecha N, Avramova Z (2010) Phosphatidylinositol 5-phosphate links dehydration stress to the activity of ARABIDOPSIS TRITHORAX-LIKE Factor ATX1. PLoS One 5:e13396. doi:10.1371/journal.pone.0013396
North HM, Almeida AD, Boutin J-P, Frey A, To A, Botran L, Sotta B, Marion-Poll A (2007) The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J 50:810–824. doi:10.1111/j.1365-313x.2007.03094.x
Ohno S (1970) Evolution by gene duplication. Springer, Berlin
Pires N, Dolan L (2010) Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol 27:862–874. doi:10.1093/molbev/msp288
Price AH, Tomos AD, Virk DS (1997) Erratum: genetic dissection of root growth in rice (Oryza sativa L.) 1: a hydroponic screen. TAG. Theor Appl Genet 95:132–142. doi:10.1007/s001220050657
Proveniers MCG, van Zanten M (2013) High temperature acclimation through PIF4 signaling. Trends Plant Sci 18:59–64. doi:10.1016/j.tplants.2012.09.002
Riboni M, Test A, Galbiati M, Tonelli C, Conti L (2014) Environmental stress and flowering time. Plant Signal Behav 9:e29036. doi:10.4161/psb.29036
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47–e47. doi:10.1093/nar/gkv007
Saleh A, Alvarez-Venegas R, Yilmaz M, Le O, Hou G, Sadder M, Al-Abdallat A, Xia Y, Lu G, Ladunga I, Avramova Z (2008) The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell Online 20:568–579. doi:10.1105/tpc.107.056614
Seiler C, Harshavardhan VT, Rajesh K, Reddy PS, Strickert M, Rolletschek H, Scholz U, Wobus U, Sreenivasulu N (2011) ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J Exp Bot 62:2615–2632. doi:10.1093/jxb/erq446
Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion-Poll A, Sun T-P, Koshiba T, Kamiya Y, Yamaguchi S, Nambara E (2006) Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 48:354–366. doi:10.1111/j.1365-313x.2006.02881.x
Seo DH, Ryu MY, Jammes F, Hwang JH, Turek M, Kang BG, Kwak JM, Kim WT (2012) Roles of four Arabidopsis U-Box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol 160:556–568. doi:10.1104/pp.112.202143
Serraj R, Krishnamurthy L, Kashiwagi J, Kumar J, Chandra S, Crouch JH (2004) Variation in root traits of chickpea (Cicer arietinum L.) grown under terminal drought. Field Crops Res 88:115–127. doi:10.1016/j.fcr.2003.12.001
Shan H, Chen S, Jiang J, Chen F, Chen Y, Gu C, Li P, Song A, Zhu X, Gao H, Zhou G, Li T, Yang X (2012) Heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance, increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana. Mol Biotech 51:160–173. doi:10.1007/s12033-011-9451-1
Sharma N, Xin R, Kim DH, Sung S, Lange T, Huq E (2016) No flowering in short day (NFL) is a bHLH transcription factor that promotes flowering specifically under short-day conditions in Arabidopsis. Development 143:682–690. doi:10.1242/dev.128595
Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T (2015) Photoperiodic flowering: time measurement mechanisms in leaves. Ann Rev Plant Biol 66:441–464. doi:10.1146/annurev-arplant-043014-115555
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K (2007) Hd3a protein is a mobile flowering signal in rice. Science 316:1033–1036. doi:10.1126/science.1141753
Toledo-Ortiz G (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell Online 15:1749–1770. doi:10.1105/tpc.013839
Tsuji H, Taoka K-I, Shimamoto K (2013) Florigen in rice: complex gene network for florigen transcription, florigen activation complex, and multiple functions. Curr Opin Plant Biol 16:228–235. doi:10.1016/j.pbi.2013.01.005
Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, Inoue H, Takehisa H, Motoyama R, Nagamura Y, Wu J, Matsumoto T, Takai T, Okuno K, Yano M (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45:1097–1102. doi:10.1038/ng.2725
Wan X-R, Li L (2006) Regulation of ABA level and water-stress tolerance of Arabidopsis by ectopic expression of a peanut 9-cis-epoxycarotenoid dioxygenase gene. Biochem Biophys Res Commun 347:1030–1038. doi:10.1016/j.bbrc.2006.07.026
Wang J, Hu J, Qian Q, Xue H-W (2013) LC2 and OsVIL2 promote rice flowering by photoperoid-induced epigenetic silencing of OsLF. Mol Plant 6:514–527. doi:10.1093/mp/sss096
Wang W-S, Zhu J, Lu Y-T (2014) Overexpression of AtbHLH112 suppresses lateral root emergence in Arabidopsis. Funct Plant Biol 41:342. doi:10.1071/fp13253
Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309:1056–1059. doi:10.1126/science.1114358
Wu H, Ye H, Yao R, Zhang T, Xiong L (2015) OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci 232:1–12. doi:10.1016/j.plantsci.2014.12.010
Xing D-H, Lai Z-B, Zheng Z-Y, Vinod KM, Fan B-F, Chen Z-X (2008) Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol Plant 1:459–470. doi:10.1093/mp/ssn020
Yoshida S, Fomo DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological studies of rice. International Rice Research Institute, Manila
Zhang C, Liu J, Zhao T, Gomez A, Li C, Yu C, Li H, Lin J, Yang Y, Liu B, Lin C (2016) A drought-inducible transcription factor delays reproductive timing in rice. Plant Physiol 171:334–343. doi:10.1104/pp.16.01691
Zhao X-L, Shi Z-Y, Peng L-T, Shen G-Z, Zhang J-L (2011) An atypical HLH protein OsLF in rice regulates flowering time and interacts with OsPIL13 and OsPIL15. New Biotechnol 28:788–797. doi:10.1016/j.nbt.2011.04.006
Zhou J, Li F, Wang J-L, Ma Y, Chong K, Xu Y-Y (2009) Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt- and osmotic stress in Arabidopsis. J Plant Physiol 166:1296–1306. doi:10.1016/j.jplph.2009.02.007
Acknowledgements
We thank Ms. Mei-Jane Fang (Live-Cell-Imaging Core Laboratory), Ms. Shu-Jen Chou (DNA Microarray Core Laboratory), and Ms. Lin-Yun Kuang (Transgenic Plant Core Facility) from the Institute of Plant and Microbial Biology at Academia Sinica, Taipei, Taiwan, for operating the confocal microscope, performing the microarray hybridization, and providing Agrobacterium-mediated stable transformation services, respectively. We also thank Mr. Shih-Hao Lee from the Crop Science Division at Taiwan Agricultural Research Institute, Taichung, Taiwan, for propagating the plant materials. We are grateful to Mr. Hau-Jiun Hsu (Department of Agronomy, National Taiwan University, Taipei, Taiwan, ROC) for analyzing the microarray data.
Funding
This work was supported by the Ministry of Science and Technology, Taipei, Taiwan (Grant No. 105-2313-B-002-044- to M.-C. Chang).
Author information
Authors and Affiliations
Contributions
HCC was responsible for the design of experiments, the data assemble and the manuscript writing. CCH carried out microarray analysis and other studies. PCL helped in conducting experiments. LYL and YWY offer great help in microarray data processing and integration. WHC and MHL were responsible for providing the experimental facilities and propagating the plant materials, respectively. MCC is the corresponding person for giving final approval of the version to be submitted. All authors have read and approved the submission of final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Hung-Chi Chen and Vicki Hsieh-Feng have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chen, HC., Hsieh-Feng, V., Liao, PC. et al. The function of OsbHLH068 is partially redundant with its homolog, AtbHLH112, in the regulation of the salt stress response but has opposite functions to control flowering in Arabidopsis . Plant Mol Biol 94, 531–548 (2017). https://doi.org/10.1007/s11103-017-0624-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-017-0624-6