Abstract
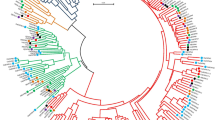
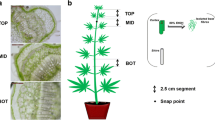
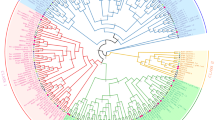
Fasciclin-like arabinogalactan proteins (FLAs), a class of arabinogalactan proteins (AGPs) are involved in plant growth and development via cell communication and adhesion. FLAs were also associated with fiber and wood formation in plants but no information is available about the roles of FLA proteins during fibre development of jute. Here, we performed molecular characterization, evolutionary relationship and expression profiling of FLAs proteins in jute (Corchorus olitorius). In total, nineteen CoFLA genes have been identified in jute genome, which were divided into four classes like FLAs of other species based on protein structure and similarity. All CoFLAs have N-terminal signal peptide and one or two FAS domain while two FLAs lack well defined AGP region and eight FLAs were devoid of C-terminal glycosylphosphatidylinositol (GPI) anchor. Expression analysis of different regions of jute stem suggested their involvement in different fiber development stages. Four genes CoFLA 11, 12, 20, and 23 were highly or predominately expressed in fiber containing bark tissues while the expression levels of six CoFLA genes 02, 03, 04, 06, 14 and 19 were comparatively higher in stick. Higher transcripts levels of CoFLA 12 and 20 in the middle bark tissues suggest their involvement in fiber elongation. In contrast, the CoFLA 11 and 23 were more expressed in bottom bark tissues suggesting their potential involvement in secondary cell wall synthesis. Our study can serve as solid foundation for further functional exploration of FLAs and in future breeding program of jute aiming fiber improvement.







Similar content being viewed by others
Data availability
The datasets supporting the conclusions and description of a complete protocol can be found within the manuscript and its additional files.
References
Faik A, Abouzouhair J, Sarhan F (2006) Putative fasciclin-like arabinogalactan-proteins (FLA) in wheat (Triticum aestivum) and rice (Oryza sativa): identification and bioinformatic analyses. Mol Genet Genom 276(5):478–494. https://doi.org/10.1007/s00438-006-0159-z
Jun L, Xiaoming W (2012) Genome-wide identification, classification and expression analysis of genes encoding putative fasciclin-like arabinogalactan proteins in Chinese cabbage (Brassica rapa L.). Mol Biol Rep 39(12):10541–10555. https://doi.org/10.1007/s11033-012-1940-1
Johnson KL, Jones BJ, Bacic A, Schultz CJ (2003) The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol 133(4):1911–1925. https://doi.org/10.1104/pp.103.031237
Showalter AM (2001) Arabinogalactan-proteins: structure, expression and function. Cell Mol Life Sci 58(10):1399–1417. https://doi.org/10.1007/pl00000784
Gaspar Y, Johnson KL, McKenna JA, Bacic A, Schultz CJ (2001) The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol Biol 47(1–2):161–176
Seifert GJ, Roberts K (2007) The biology of arabinogalactan proteins. Annu Rev Plant Biol 58(1):137–161. https://doi.org/10.1146/annurev.arplant.58.032806.103801
Pereira AM, Pereira LG, Coimbra S (2015) Arabinogalactan proteins: rising attention from plant biologists. Plant Reprod 28(1):1–15. https://doi.org/10.1007/s00497-015-0254-6
Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A (2002) Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiol 129(4):1448–1463. https://doi.org/10.1104/pp.003459
Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR (2010) A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol 153(2):485–513. https://doi.org/10.1104/pp.110.156554
Zang L, Zheng T, Chu Y, Ding C, Zhang W, Huang Q, Su X (2015) Genome-wide analysis of the fasciclin-like arabinogalactan protein gene family reveals differential expression patterns, localization, and salt stress response in populus. Front Plant Sci 6:1140. https://doi.org/10.3389/fpls.2015.01140
Elkins T, Zinn K, McAllister L, Hoffmann FM, Goodman CS (1990) Genetic analysis of a drosophila neural cell adhesion molecule: interaction of fasciclin I and Abelson tyrosine kinase mutations. Cell 60(4):565–575. https://doi.org/10.1016/0092-8674(90)90660-7
Huber O, Sumper M (1994) Algal-CAMs: isoforms of a cell adhesion molecule in embryos of the alga Volvox with homology to drosophila fasciclin I. EMBO J 13(18):4212–4222
Kawamoto T, Noshiro M, Shen M, Nakamasu K, Hashimoto K, Kawashima-Ohya Y, Gotoh O, Kato Y (1998) Structural and phylogenetic analyses of RGD-CAP/beta ig-h3, a fasciclin-like adhesion protein expressed in chick chondrocytes. Biochim Biophys Acta 1395(3):288–292. https://doi.org/10.1016/s0167-4781(97)00172-3
Tan L, Leykam JF, Kieliszewski MJ (2003) Glycosylation motifs that direct arabinogalactan addition to arabinogalactan-proteins. Plant Physiol 132(3):1362–1369. https://doi.org/10.1104/pp.103.021766
Huang GQ, Xu WL, Gong SY, Li B, Wang XL, Xu D, Li XB (2008) Characterization of 19 novel cotton FLA genes and their expression profiling in fiber development and in response to phytohormones and salt stress. Physiol Plant 134(2):348–359. https://doi.org/10.1111/j.1399-3054.2008.01139.x
Guerriero G, Mangeot-Peter L, Legay S, Behr M, Lutts S, Siddiqui KS, Hausman JF (2017) Identification of fasciclin-like arabinogalactan proteins in textile hemp (Cannabis sativa L.): in silico analyses and gene expression patterns in different tissues. BMC Genom 18(1):741. https://doi.org/10.1186/s12864-017-3970-5
MacMillan CP, Taylor L, Bi Y, Southerton SG, Evans R, Spokevicius A (2015) The fasciclin-like arabinogalactan protein family of Eucalyptus grandis contains members that impact wood biology and biomechanics. New Phytol 206(4):1314–1327. https://doi.org/10.1111/nph.13320
Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK (2003) The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15(1):19–32. https://doi.org/10.1105/tpc.007872
Johnson KL, Kibble NA, Bacic A, Schultz CJ (2011) A fasciclin-like arabinogalactan-protein (FLA) mutant of Arabidopsis thaliana, fla1, shows defects in shoot regeneration. PLoS ONE 6(9):e25154. https://doi.org/10.1371/journal.pone.0025154
MacMillan CP, Mansfield SD, Stachurski ZH, Evans R, Southerton SG (2010) Fasciclin-like arabinogalactan proteins: specialization for stem biomechanics and cell wall architecture in Arabidopsis and Eucalyptus. Plant J 62(4):689–703. https://doi.org/10.1111/j.1365-313X.2010.04181.x
Wang H, Jiang C, Wang C, Yang Y, Yang L, Gao X, Zhang H (2015) Antisense expression of the fasciclin-like arabinogalactan protein FLA6 gene in populus inhibits expression of its homologous genes and alters stem biomechanics and cell wall composition in transgenic trees. J Exp Bot 66(5):1291–1302. https://doi.org/10.1093/jxb/eru479
Huang GQ, Gong SY, Xu WL, Li W, Li P, Zhang CJ, Li DD, Zheng Y, Li FG, Li XB (2013) A fasciclin-like arabinogalactan protein, GhFLA1, is involved in fiber initiation and elongation of cotton. Plant Physiol 161(3):1278–1290. https://doi.org/10.1104/pp.112.203760
Roach MJ, Deyholos MK (2007) Microarray analysis of flax (Linum usitatissimum L.) stems identifies transcripts enriched in fibre-bearing phloem tissues. Mol Genet Genom 278(2):149–165. https://doi.org/10.1007/s00438-007-0241-1
Roach MJ, Deyholos MK (2008) Microarray analysis of developing flax hypocotyls identifies novel transcripts correlated with specific stages of phloem fibre differentiation. Ann Bot 102(3):317–330. https://doi.org/10.1093/aob/mcn110
Gorshkova T, Chernova T, Mokshina N, Gorshkov V, Kozlova L, Gorshkov O (2018) Transcriptome analysis of intrusively growing flax fibers isolated by laser microdissection. Sci Rep 8(1):14570. https://doi.org/10.1038/s41598-018-32869-2
Townsend T, Sette J (2016) Natural Fibres and the World Economy. In: Fangueiro R, Rana S (eds) Natural fibres: advances in science and technology towards industrial applications. RILEM bookseries, vol 12. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-7515-1_30
Shafrin F, Ferdous AS, Sarkar SK, Ahmed R, Amin A, Hossain K, Sarker M, Rencoret J, Gutierrez A, Del Rio JC, Sanan-Mishra N, Khan H (2017) Modification of monolignol biosynthetic pathway in jute: different gene, different consequence. Sci Rep 7:39984. https://doi.org/10.1038/srep39984
Choudhary SB, Kumar M, Chowdhury I, Singh RK, Pandey SP, Sharma HK, Karmakar PG (2016) An efficient and cost effective method of RNA extraction from mucilage, phenol and secondary metabolite rich bark tissue of tossa jute (C. olitorius L.) actively developing phloem fiber. 3 Biotech 6(1):100. https://doi.org/10.1007/s13205-016-0415-9
Tanmoy AM, Alum AM, Islam MS, Farzana T, Khan H (2015) Jute (Corchorus olitorius var. O-72) stem lignin: variation in content with age. Bangladesh J Bot 43(3):309–314. https://doi.org/10.3329/bjb.v43i3.21603
del Rio JC, Rencoret J, Marques G, Li J, Gellerstedt G, Jimenez-Barbero J, Martinez AT, Gutierrez A (2009) Structural characterization of the lignin from jute (Corchorus capsularis) fibers. J Agric Food Chem 57(21):10271–10281. https://doi.org/10.1021/jf900815x
Islam MS, Saito JA, Emdad EM, Ahmed B, Islam MM, Halim A, Hossen QM, Hossain MZ, Ahmed R, Hossain MS, Kabir SM, Khan MS, Khan MM, Hasan R, Aktar N, Honi U, Islam R, Rashid MM, Wan X, Hou S, Haque T, Azam MS, Moosa MM, Elias SM, Hasan AM, Mahmood N, Shafiuddin M, Shahid S, Shommu NS, Jahan S, Roy S, Chowdhury A, Akhand AI, Nisho GM, Uddin KS, Rabeya T, Hoque SM, Snigdha AR, Mortoza S, Matin SA, Islam MK, Lashkar MZ, Zaman M, Yuryev A, Uddin MK, Rahman MS, Haque MS, Alam MM, Khan H, Alam M (2017) Comparative genomics of two jute species and insight into fibre biogenesis. Nat Plants 3:16223. https://doi.org/10.1038/nplants.2016.223
Chakraborty A, Sarkar D, Satya P, Karmakar PG, Singh NK (2015) Pathways associated with lignin biosynthesis in lignomaniac jute fibres. Mol Genet Genom 290(4):1523–1542. https://doi.org/10.1007/s00438-015-1013-y
Zhang L, Ming R, Zhang J, Tao A, Fang P, Qi J (2015) De novo transcriptome sequence and identification of major bast-related genes involved in cellulose biosynthesis in jute (Corchorus capsularis L.). BMC Genom 16:1062. https://doi.org/10.1186/s12864-015-2256-z
Gorshkova T, Sal'nikov V, Chemikosova S, Ageeva M, Pavlencheva N, Dam J (2003) The snap point: a transition point in Linum usitatissimum bast fiber development. Ind Crops Prod 18:213–221. https://doi.org/10.1016/S0926-6690(03)00043-8
Esau K (1943) Vascular differentiation in the vegetative shoots of Linum III. The origin of the bast fibres. Am J Bot 30:579–586
Ageeva MV, Petrovska B, Kieft H, Salnikov VV, Snegireva AV, van Dam JEG, van Veenendaal WLH, Emons AMC, Gorshkova TA, van Lammeren AAM (2005) Intrusive growth of flax phloem fibers is of intercalary type. Planta 222:565–574
Gorshkova TA, Wyatt SE, Salnikov VV, Gibeaut DM, Ibragimov MR, Lozovaya VV, Carpita NC (1996) Cell-wall polysaccharides of developing flax plants. Plant Physiol 110:721–729
Gorshkova TA, Salnikov VV, Pogodina NM, Chemikosova SB, Yablokova EV, Ulanov AV, Ageeva MV, van Dam JEG, Lozovaya VV (2000) Composition and distribution of cell wall phenolic compounds in flax (Linum usitatissimum L.) stem tissues. Ann Bot 85:477–486
Day A, Ruel K, Neutelings G, Cronier D, David H, Hawkins S, Chabbert B (2005) Lignification in the flax stem: evidence for an unusual lignin in bast fibers. Planta 222:234–245
Esau K (1977) Anatomy of seed plants, 2nd edn. Wiley, London
McDougall GJ (1991) Cell wall-associated peroxidases and lignification during growth of flax fibres. J Plant Physiol 39:182–186
Kundu A, Sarkar D, Mandal NA, Sinha MK, Bs M (2011) A secondary phloic (bast) fibre-shy (bfs) mutant of dark jute (Corchorus olitorius L.) develops lignified fibre cells but is defective in cambial activity. Plant Growth Regul 67:45–55
Eddy SR (1998) Profile hidden Markov models. Bioinformatics (Oxford, England) 14(9):755–763. https://doi.org/10.1093/bioinformatics/14.9.755
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40:D302–305. https://doi.org/10.1093/nar/gkr931
Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–251. https://doi.org/10.1093/nar/gkj149
Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H (2019) SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37(4):420–423. https://doi.org/10.1038/s41587-019-0036-z
Eisenhaber B, Wildpaner M, Schultz CJ, Borner GH, Dupree P, Eisenhaber F (2003) Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiol 133(4):1691–1701. https://doi.org/10.1104/pp.103.023580
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H (2012) ExPASy: SIB bioinformatics resource portal. Nucleic acids Res 40:W597–603. https://doi.org/10.1093/nar/gks400
Chou KC, Shen HB (2010) Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 5(6):e11335. https://doi.org/10.1371/journal.pone.0011335
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics (Oxford, England) 31(8):1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36
Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36(10):3420–3435. https://doi.org/10.1093/nar/gkn176
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327. https://doi.org/10.1093/nar/30.1.325
Koziel SP (2010). Genetic analysis of lignification and secondary wall development in bast fibers of industrial Hemp (Cannabis sativa). Master’s Thesis, University of Alberta, Edmonton, AB, Canada
Ahmed R, Hossain MS, Haque M, Alam M, Islam MS (2019) Modified protocol for RNA isolation from different parts of field-grown jute plant suitable for NGS data generation and quantitative real-time RT-PCR. Afr J Biotechnol 18:647–658. https://doi.org/10.5897/AJB2019.16819
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods (San Diego, Calif) 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Hossain MS, Ahmed R, Haque MS, Alam MM, Islam MS (2019) Identification and validation of reference genes for real-time quantitative RT-PCR analysis in jute. BMC Mol Biol 20(1):13. https://doi.org/10.1186/s12867-019-0130-2
Kim JE, Kim SJ, Lee BH, Park RW, Kim KS, Kim IS (2000) Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-beta-induced gene, betaig-h3. J Biol Chem 275(40):30907–30915. https://doi.org/10.1074/jbc.M002752200
Hobson N, Deyholos MK (2013) LuFLA1PRO and LuBGAL1PRO promote gene expression in the phloem fibres of flax (Linum usitatissimum). Plant Cell Rep 32:517–528. https://doi.org/10.1007/s00299-013-1383-8
Mokshina N, Chernova T, Galinousky D, Gorshkov O, Gorshkova T (2018) Key stages of fiber development as determinants of bast fiber yield and quality. Fibers 6:20. https://doi.org/10.3390/fib6020020
Snegireva A, Chernova T, Ageeva M, Lev-Yadun S, Gorshkova T (2015) Intrusive growth of primary and secondary phloem fibres in hemp stem determines fibre-bundle formation and structure. AoB Plants. https://doi.org/10.1093/aobpla/plv061
Ito S, Suzuki Y, Miyamoto K, Ueda J, Yamaguchi I (2005) AtFLA11, a fasciclin-like arabinogalactan-protein, specifically localized in sclerenchyma cells. Biosci Biotechnol Biochem 69(10):1963–1969. https://doi.org/10.1271/bbb.69.1963
Acknowledgements
The authors are thankful to Emdadul Mannan Emdad for his technical support during bioinformatics analysis.
Funding
The author(s) received no specific funding for this work.
Author information
Authors and Affiliations
Contributions
MSI and MS Hossain designed the research. MS Hossain carried out the experiments, analyzed the data and wrote the manuscript. BA and MS Hossain contributed in bioinformatics analysis. MWU and NA revised the manuscript. MS Haque and MSI directed and reviewed the manuscript. All the authors read, discussed and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hossain, M.S., Ahmed, B., Ullah, M.W. et al. Genome-wide identification of fasciclin-like arabinogalactan proteins in jute and their expression pattern during fiber formation. Mol Biol Rep 47, 7815–7829 (2020). https://doi.org/10.1007/s11033-020-05858-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05858-w