Abstract
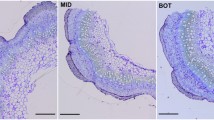
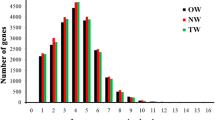
To better understand the molecular processes associated with the development of the unusually long (>30 mm) and strong bast fibre cells within the phloem of flax stems, we conducted a gene discovery experiment to identify transcripts enriched in fibre-bearing tissues, with the intention that these transcripts would serve as future targets for crop improvement and research in phloem development and cell wall deposition. We produced a library of 9,600 cDNA clones from the peels of flax stems, and selected tissue-specific cDNAs for sequencing based on two series of microarray experiments. In the first microarray series, we compared transcript abundance in stem-peels and leaves, and identified stem-enriched transcripts putatively involved in the processes of polysaccharide and cell wall metabolism. In the second microarray series, we compared gene expression in three segments of the vertical stem axis, which constituted a developmental series for phloem fibres and other cell types. The expression of specific LTP and AGP transcripts was particularly well-correlated with stem segments during either the elongation phase or cell-wall thickening phase of phloem fibre development, and the phloem-specific enrichment of these transcripts was confirmed by qRT-PCR. Transcripts representing multiple, distinct chitinases, β-galactosidases, arabinogalactan proteins (AGP), and lipid transfer proteins (LTPs) were among the interesting transcripts enriched in specific stages of the developing stem. Considered together, the results of our analyses suggest similarity between the molecular mechanisms underlying phloem fibre development and the gelatinous fibres of tension wood in trees.








Similar content being viewed by others
References
Ageeva M et al (2005) Intrusive growth of flax phloem fibers is of intercalary type. Planta 222:565–574
Altschul SF et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Andeme-Onzighi C, Girault R, His I, Morvan C, Driouich A (2000) Immunocytochemical characterization of early-developing flax fiber cell walls. Protoplasma 213:235–245
Andeme-Onzighi C, Sivaguru M, Judy-March J, Baskin TI, Driouich A (2002) The reb1–1 mutation of Arabidopsis alters the morphology of trichoblasts, the expression of arabinogalactan proteins, and the organization of cortical microtubules. Planta 215:949–958
Andersson-Gunneras S et al (2006) Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J 45:144–165
Ben-Naim O et al (2006) The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J 46:462–476
Bennett MD, Smith JB (1976) Nuclear DNA amounts in angiosperms. Philos Trans R Soc Lond B 274:227–274
Biddulph O, Cory R (1965) Translocation of C14 metabolites in Phloem of bean plant. Plant Physiol 40:119
Blee K et al (2001) Proteomic analysis reveals a novel set of cell wall proteins in a transformed tobacco cell culture that synthesises secondary walls as determined by biochemical and morphological parameters. Planta 212:404–415
Bolle C (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218:683–692
Burn JE, Hocart CH, Birch RJ, Cork AC, Williamson RE (2002) Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol 129:797–807
Bush MS, Marry M, Huxham IM, Jarvis MC, McCann MC (2001) Developmental regulation of pectic epitopes during potato tuberisation. Planta 213:869–880
Chaumont F, Barrieu F, Herman EM, Chrispeels MJ (1998) Characterization of a maize tonoplast aquaporin expressed in zones of cell division and elongation. Plant Physiol 117:1143–1152
Chemikosova SB, Pavlencheva NV, Gur’yanov OP, Gorshkova TA (2006) The effect of soil drought on the phloem fiber development in long-fiber flax. Russ J Plant Physiol 53:656–662
Chen LM, Carpita NC, Reiter WD, Wilson RH, Jeffries C, McCann MC (1998) A rapid method to screen for cell-wall mutants using discriminant analysis of Fourier transform infrared spectra. Plant J 16:385–392
Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6:850–861
Day A et al (2005a) ESTs from the fibre-bearing stem tissues of flax (Linum usitatissimum L.): expression analyses of sequences related to cell wall development. Plant Biol 7:23–32
Day A et al (2005b) Lignification in the flax stem: evidence for an unusual lignin in bast fibers. Planta 222:234–245
Demura T, Fukuda H (1993) Molecular cloning and characterization of cDNAs associated with tracheary element differentiation in cultured Zinnia cells. Plant Physiol 103:815–821
Doblin MS, Kurek I, Jacob-Wilk D, Delmer DP (2002) Cellulose biosynthesis in plants: from genes to rosettes. Plant Cell Physiol 43:1407–1420
Edwards D, Murray J, Smith A (1998) Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol 117:1015–1022
Ehlting J et al (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42:618–640
Esau K (1943) Vascular differentiation in the vegetative shoot of Linum III. The origin of the bast fibers. Am J Bot 30:579–586
Ewing B, Hillier L, Wendl MC, Green P (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8:175–185
Farrokhi N et al (2006) Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol J 4:145–167
Favery B et al (2001) KOJAK encodes a cellulose synthase-like protein required for root hair cell morphogenesis in Arabidopsis. Genes Dev 15:79–89
Franck RR (2005) Overview. In: Franck RR (ed) Bast and other plant fibres. CRC Press, New York, pp 1–23
Fukuda H (2004) Signals that control plant vascular cell differentiation. Nat Rev Mol Cell Biol 5:379–391
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Gorshkova TA et al (2004) Occurrence of cell-specific galactan is coinciding with bast fiber developmental transition in flax. Ind Crops Prod 19:217–224
Gorshkova TA, Morvan C (2006) Secondary cell-wall assembly in flax phloem fibres: role of galactans. Planta 223:149–158
Gorshkova TA, Sal’nikova VV, Chemikosova SB, Ageeva MV, Pavlencheva NV, van Dam JEG (2003) The snap point: a transition point in Linum usitatissimum bast fiber development. Ind Crops Prod 18:213–221
Gorshkova TA et al (2000) Composition and distribution of cell wall phenolic compounds in flax (Linum usitatissimum L.) stem tissues. Ann Bot 85:477–486
Haigler CH et al (2001) Carbon partitioning to cellulose synthesis. Plant Mol Biol 47:29–51
His I, Andeme-Onzighi C, Morvan C, Driouich A (2001) Microscopic studies on mature flax fibers embedded in LR white: immunogold localization of cell wall matrix polysaccharicles. J Histochem Cytochem 49:1525–1535
Iseli C, Jongeneel C, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol, pp 138–148
Johnson KL, Jones BJ, Bacic A, Schultz CJ (2003) The fasciclin-like arabinogalactan proteins of arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol 133:1911–1925
Joseleau JP, Imai T, Kuroda K, Ruel K (2004) Detection in situ and characterization of lignin in the G-layer of tension wood fibres of Populus deltoides. Planta 219:338–345
Karkonen A, Fry SC (2006) Effect of ascorbate and its oxidation products on H2O2 production in cell-suspension cultures of Picea abies and in the absence of cells. J Exp Bot 57:1633–1644
Ko TS, Lee SM, Schaefer SC, Korban SS (2003) Characterization of a tissue-specific and developmentally regulated beta-1,3-glucanase gene family in Prunus persica. Plant Physiol Biochem 41:955–963
Kulczyk A, Yang J, Neuhaus D (2004) Solution structure and DNA binding of the zinc-finger domain from DNA ligase III alpha. J Mol Biol 341:723–738
Lafarguette F et al (2004) Poplar genes encoding fasciclin-like arabinogalactan proteins are highly expressed in tension wood. New Phytol 164:107–121
Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci 103:6055–6060
Lee KJD et al (2005) Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. Plant Cell 17:3051–3065
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25:402–408
Lorkovic Z, Barta A (2002) Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res 30:623–635
Martin I et al (2005) In vivo expression of a Cicer arietinum beta-galactosidase in potato tubers leads to a reduction of the galactan side-chains in cell wall pectin. Plant Cell Physiol 46:1613–1622
McCartney L, Ormerod AP, Gidley MJ, Knox JP (2000) Temporal and spatial regulation of pectic (1→4)-beta-D-galactan in cell walls of developing pea cotyledons: implications for mechanical properties. Plant J 22:105–113
McCartney L, Steele-King CG, Jordan E, Knox JP (2003) Cell wall pectic (1→4)-beta-D-galactan marks the acceleration of cell elongation in the Arabidopsis seedling root meristem. Plant J 33:447–454
Minic Z, Jouanin L (2006) Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiol Biochem 44:435–449
Mooney C, Stolle-Smits T, Schols H, de Jong E (2001) Analysis of retted and non retted flax fibres by chemical and enzymatic means. J Biotechnol 89:205–216
Motose H, Sugiyama M, Fukuda H (2004) A proteoglycan mediates inductive interaction during plant vascular development. Nature 429:873–878
Mouille G, Robin S, Lecomte M, Pagant S, Hofte H (2003) Classification and identification of Arabidopsis cell wall mutants using Fourier-transform infraRed (FT-IR) microspectroscopy. Plant J 35:393–404
Namasivayam P, Baldwin TC (1997) Tissue print analyses of soluble cell wall components present in developing stems of Carica papaya (Papaya). Asia Pac J Mol Biol Biotechnol 5:165–172
Nessler CL, Kurz WGW, Pelcher LE (1990) Isolation and analysis of the major latex protein genes of opium poppy. Plant Mol Biol 15:951–953
Nieuwland J et al (2005) Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell 17:2009–2019
Ohashi-Ito K, Kubo M, Demura T, Fukuda H (2005) Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol 46:1646–1656
Orford SJ, Timmis JN (2000) Expression of a lipid transfer protein gene family during cotton fibre development. Biochim Biophys Acta Mol Cell Biol Lipids 1483:275–284
Parkinson H et al (2007) ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res 35:D747–D750
Paux E et al (2005) Transcript profiling of Eucalyptus xylem genes during tension wood formation. New Phytol 167:89–100
Prassinos C, Ko JH, Han KH (2005) Transcriptome profiling of vertical stem segments provides insights into the genetic regulation of secondary growth in hybrid aspen trees. Plant Cell Physiol 46:1213–1225
Rinne P, van der Schoot C (1998) Symplasmic fields in the tunica of the shoot apical meristem coordinate morphogenetic events. Development 125:1477–1485
Roger D, Gallusci P, Meyer Y, David A, David H (1998) Basic chitinases are correlated with the morphogenic response of flax cells. Physiol Plant 103:271–279
Ruan YL, Xu SM, White R, Furbank RT (2004) Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fiber elongation mediated by callose turnover. Plant Physiol 136:4104–4113
Saeed AI et al (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378
Sarria R et al (2001) Characterization of a family of arabidopsis genes related to xyloglucan fucosyltransferase1. Plant Physiol 127:1595–1606
Serpe MD, Muir AJ, Keidel AM (2001) Localization of cell wall polysaccharides in nonarticulated laticifers of Asclepias speciosa Torr. Protoplasma 216:215–226
Sieburth LE, Deyholos MK (2006) Vascular development: the long and winding road. Curr Opin Plant Biol 9:48–54
Seifert GJ, Barber C, Wells B, Roberts K (2004) Growth regulators and the control of nucleotide sugar flux. Plant Cell 16:723–730
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4:447–456
Taguchi T, Uraguchi A, Katsumi M (1999) Auxin- and acid-induced changes in the mechanical properties of the cell wall. Plant Cell Physiol 40:743–749
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98:5116–5121
Vicre M, Jauneau A, Knox JP, Driouich A (1998) Immunolocalization of beta-(1→4) and beta-(1→6)-D-galactan epitopes in the cell wall and Golgi stacks of developing flax root tissues. Protoplasma 203:26–34
Vilaine F, Palauqui JC, Amselem J, Kusiak C, Lemoine R, Dinant S (2003) Towards deciphering phloem: a transcriptome analysis of the phloem of Apium graveolens. Plant J 36:67–81
Zhang DS et al (2004) Members of a new group of chitinase-like genes are expressed preferentially in cotton cells with secondary walls. Plant Mol Biol 54:353–372
Zhao CS, Craig JC, Petzold HE, Dickerman AW, Beers EP (2005) The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol 138:803–818
Acknowledgments
We thank Drs. Larry Pelcher (Plant Biotechnology Institute, Saskatoon, Canada) and Anthony Cornish (University of Alberta), for assistance with sequencing and microarray preparation, respectively. We also thank Charlie Hsu for assistance in preparation of the microarray amplicons. This research is funded by Alberta Innovation and Science, and by the Natural Sciences and Engineering Research Council (Canada).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Tyagi.
Nucleotide sequence data reported are available in the GenBank database under the accession numbers EH791145-EH792584, and EL585326-EL585347. Microarray data are available in the ArrayExpress database as accessions E-MEXP-989 and E-MEXP-990.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Roach, M.J., Deyholos, M.K. Microarray analysis of flax (Linum usitatissimum L.) stems identifies transcripts enriched in fibre-bearing phloem tissues. Mol Genet Genomics 278, 149–165 (2007). https://doi.org/10.1007/s00438-007-0241-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-007-0241-1