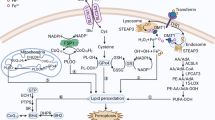
Abstract
Huntington’s disease (HD) is an inherited, autosomal, neurodegenerative ailment that affects the striatum of the brain. Despite its debilitating effect on its patients, there is no proven cure for HD management as of yet. Neuroinflammation, excitotoxicity, and environmental factors have been reported to influence the regulation of gene expression by modifying epigenetic mechanisms. Aside focusing on the etiology, changes in epigenetic mechanisms have become a crucial factor influencing the interaction between HTT protein and epigenetically transcribed genes involved in neuroinflammation and HD. This review presents relevant literature on epigenetics with special emphasis on neuroinflammation and HD. It summarizes pertinent research on the role of neuroinflammation and post-translational modifications of chromatin, including DNA methylation, histone modification, and miRNAs. To achieve this about 1500 articles were reviewed via databases like PubMed, ScienceDirect, Google Scholar, and Web of Science. They were reduced to 534 using MeSH words like ‘epigenetics, neuroinflammation, and HD’ coupled with Boolean operators. Results indicated that major contributing factors to the development of HD such as mitochondrial dysfunction, excitotoxicity, neuroinflammation, and apoptosis are affected by epigenetic alterations. However, the association between neuroinflammation-altered epigenetics and the reported transcriptional changes in HD is unknown. Also, the link between epigenetically dysregulated genomic regions and specific DNA sequences suggests the likelihood that transcription factors, chromatin-remodeling proteins, and enzymes that affect gene expression are all disrupted simultaneously. Hence, therapies that target pathogenic pathways in HD, including neuroinflammation, transcriptional dysregulation, triplet instability, vesicle trafficking dysfunction, and protein degradation, need to be developed.



Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- HD:
-
Huntington’s disease
- HTT:
-
Huntingtin gene
- mHtt:
-
Mutant huntingtin protein
- PPAR-GC1α:
-
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PTM:
-
Post-translational modifications
- DNMTs:
-
DNA methyltransferase enzymes
- TET:
-
Ten-eleven translocation
- 7-mG:
-
7-Methyl guanine
- HPLC–UV:
-
High-performance liquid chromatography–ultraviolet spectroscopy
- LC–MS/MS:
-
Liquid chromatography coupled with tandem mass spectrometry
- RFLP:
-
Restriction fragment length polymorphism
- AFLP:
-
Amplification fragment length polymorphism
- HAT:
-
Histone acetyltransferase
- HDAC:
-
Histone deacetylase
- HMT:
-
Histone methyltransferase
- ESET/SETDB1:
-
ERG-associated protein with SET domain
- REST:
-
Repressor element 1 (RE1) silencing transcription factor
- PRMT5:
-
Arginine dimethyltransferase 5
- CREB:
-
cAMP response element binding
References
Abdanipour A, Schluesener HJ, Tiraihi T (2012) Effects of valproic acid, a histone deacetylase inhibitor, on improvement of locomotor function in rat spinal cord injury based on epigenetic science. Iran Biomed J 16(2):90–100
Arthur R, Navik U, Kumar P (2022) Repurposing artemisinins as neuroprotective agents: a focus on the PI3k/Akt signalling pathway. Naunyn Schmiedebergs Arch Pharmacol 396(4):593–605
Athira KV, Sadanandan P, Chakravarty S (2021) Repurposing vorinostat for the treatment of disorders affecting brain. Neuromolecular Med 23(4):449–465
Ban JJ et al (2017) MicroRNA-27a reduces mutant huntingtin aggregation in an in vitro model of Huntington’s disease. Biochem Biophys Res Commun 488(2):316–321
Bassi S et al (2017) Epigenetics of Huntington’s disease. Adv Exp Med Biol 978:277–299
Berson A et al (2018) Epigenetic regulation in neurodegenerative diseases. Trends Neurosci 41(9):587–598
Biagioli M et al (2015) Htt CAG repeat expansion confers pleiotropic gains of mutant huntingtin function in chromatin regulation. Hum Mol Genet 24(9):2442–2457
Breiling A, Lyko F (2015) Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin 8:24
Campos EI, Reinberg D (2009) Histones: annotating chromatin. Annu Rev Genet 43:559–599
Caron NS, Dorsey ER, Hayden MR (2018) Therapeutic approaches to Huntington disease: from the bench to the clinic. Nat Rev Drug Discov 17(10):729–750
Chang KH, Wu YR, Chen CM (2017) Down-regulation of miR-9* in the peripheral leukocytes of Huntington’s disease patients. Orphanet J Rare Dis 12(1):185
Cheng PH et al (2013) miR-196a ameliorates phenotypes of Huntington disease in cell, transgenic mouse, and induced pluripotent stem cell models. Am J Hum Genet 93(2):306–312
Cheray M, Joseph B (2018) Epigenetics control microglia plasticity. Front Cell Neurosci 12:243
Chouliaras L et al (2010) Epigenetic regulation in the pathophysiology of Alzheimer’s disease. Prog Neurobiol 90(4):498–510
Das S, Bhattacharyya NP (2015) Heat shock factor 1-regulated miRNAs can target huntingtin and suppress aggregates of mutant huntingtin. Microrna 4(3):185–193
Das E, Jana NR, Bhattacharyya NP (2015) Delayed cell cycle progression in STHdh(Q111)/Hdh(Q111) cells, a cell model for Huntington’s disease mediated by microRNA-19a, microRNA-146a and microRNA-432. Microrna 4(2):86–100
De Souza RA et al (2016) DNA methylation profiling in human Huntington’s disease brain. Hum Mol Genet 25(10):2013–2030
Díez-Planelles C et al (2016) Circulating microRNAs in Huntington’s disease: emerging mediators in metabolic impairment. Pharmacol Res 108:102–110
Dompierre JP et al (2007) Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J Neurosci 27(13):3571–3583
Dong X, Cong S (2021) MicroRNAs in Huntington’s disease: diagnostic biomarkers or therapeutic agents? Front Cell Neurosci 15:705348
Ebbel EN et al (2010) Identification of phenylbutyrate-generated metabolites in Huntington disease patients using parallel liquid chromatography/electrochemical array/mass spectrometry and off-line tandem mass spectrometry. Anal Biochem 399(2):152–161
Faragó A et al (2022) Acetylation state of lysine 14 of histone H33 affects mutant huntingtin induced pathogenesis. Int J Mol Sci 23(23):15173
Feng J et al (2010) Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci 13(4):423–430
Ferrante RJ et al (2004) Chemotherapy for the brain: the antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington’s disease. J Neurosci 24(46):10335–10342
Gardian G et al (2005) Neuroprotective effects of phenylbutyrate in the N171–82Q transgenic mouse model of Huntington’s disease. J Biol Chem 280(1):556–563
Ghosh P, Saadat A (2021) Neurodegeneration and epigenetics: a review. Neurologia (Engl Ed)
Ghosh R, Tabrizi SJ (2018) Clinical features of Huntington’s disease. Adv Exp Med Biol 1049:1–28
Glajch KE, Sadri-Vakili G (2015) Epigenetic mechanisms involved in Huntington’s disease pathogenesis. J Huntingtons Dis 4(1):1–15
Gray SG (2010) Targeting histone deacetylases for the treatment of Huntington’s disease. CNS Neurosci Ther 16(6):348–361
Guedes-Dias P et al (2015) HDAC6 inhibition induces mitochondrial fusion, autophagic flux and reduces diffuse mutant huntingtin in striatal neurons. Biochim Biophys Acta 1852(11):2484–2493
Guo JU et al (2011) Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci 14(10):1345–1351
Her LS et al (2017) miR-196a enhances neuronal morphology through suppressing RANBP10 to provide neuroprotection in Huntington’s disease. Theranostics 7(9):2452–2462
Horvath S et al (2016) Huntington’s disease accelerates epigenetic aging of human brain and disrupts DNA methylation levels. Aging (Albany NY) 8(7):1485–1512
Hur K et al (2014) Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 63(4):635–646
Hyeon JW, Kim AH, Yano H (2021) Epigenetic regulation in Huntington’s disease. Neurochem Int 148:105074
Igarashi S et al (2003) Inducible PC12 cell model of Huntington’s disease shows toxicity and decreased histone acetylation. NeuroReport 14(4):565–568
Islam MT (2017) Oxidative stress and mitochondrial dysfunction-linked neurodegenerative disorders. Neurol Res 39(1):73–82
Jia H et al (2015) HDAC inhibition imparts beneficial transgenerational effects in Huntington’s disease mice via altered DNA and histone methylation. Proc Natl Acad Sci U S A 112(1):E56–E64
Jia H et al (2016) The effects of pharmacological inhibition of histone deacetylase 3 (HDAC3) in Huntington’s disease mice. PLoS One 11(3):e0152498
Jimenez-Sanchez M et al (2017) Huntington’s disease: mechanisms of pathogenesis and therapeutic strategies. Cold Spring Harb Perspect Med 7(7):a024240
Johnson EB et al (2021) Dynamics of cortical degeneration over a decade in Huntington’s disease. Biol Psychiatry 89(8):807–816
Jovicic A et al (2013) MicroRNA-22 (miR-22) overexpression is neuroprotective via general anti-apoptotic effects and may also target specific Huntington’s disease-related mechanisms. PLoS One 8(1):e54222
Jurcau A (2022) Molecular pathophysiological mechanisms in Huntington’s disease. Biomedicines 10(6):1432
Juźwik CA et al (2019) microRNA dysregulation in neurodegenerative diseases: a systematic review. Prog Neurobiol 182:101664
Kanherkar RR, Bhatia-Dey N, Csoka AB (2014) Epigenetics across the human lifespan. Front Cell Dev Biol 2:49
Kaur G et al (2022) DNA methylation: a promising approach in management of Alzheimer’s disease and other neurodegenerative disorders. Biology (Basel) 11(1):90
Kawahara Y (2014) Human diseases caused by germline and somatic abnormalities in microRNA and microRNA-related genes. Congenit Anom (Kyoto) 54(1):12–21
Killoran A et al (2013) Characterization of the Huntington intermediate CAG repeat expansion phenotype in PHAROS. Neurology 80(22):2022–2027
Kim A et al (2021) New avenues for the treatment of Huntington’s disease. Int J Mol Sci 22(16):8363
Kimura H (2013) Histone modifications for human epigenome analysis. J Hum Genet 58(7):439–445
Kumar V et al (2022) Understanding the role of histone deacetylase and their inhibitors in neurodegenerative disorders: current targets and future perspective. Curr Neuropharmacol 20(1):158–178
Kunkanjanawan T et al (2016) miR-196a ameliorates cytotoxicity and cellular phenotype in transgenic Huntington’s disease monkey neural cells. PLoS One 11(9):e0162788
Kurdyukov S, Bullock M (2016) DNA methylation analysis: choosing the right method. Biology (Basel) 5(1):3
Laird PW (2010) Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet 11(3):191–203
Le Gras S et al (2017) Altered enhancer transcription underlies Huntington’s disease striatal transcriptional signature. Sci Rep 7:42875
Le Martelot G et al (2012) Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol 10(11):e1001442
Lee J et al (2008) Monoallele deletion of CBP leads to pericentromeric heterochromatin condensation through ESET expression and histone H3 (K9) methylation. Hum Mol Genet 17(12):1774–1782
Lee J et al (2013) Epigenetic mechanisms of neurodegeneration in Huntington’s disease. Neurotherapeutics 10(4):664–676
Liddelow SA et al (2017) Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541(7638):481–487
Love IM et al (2012) The histone acetyltransferase PCAF regulates p21 transcription through stress-induced acetylation of histone H3. Cell Cycle 11(13):2458–2466
Lu AT et al (2020) DNA methylation study of Huntington’s disease and motor progression in patients and in animal models. Nat Commun 11(1):4529
Marques SC et al (2010) Alzheimer’s disease: the quest to understand complexity. J Alzheimers Dis 21(2):373–383
Martí E et al (2010) A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res 38(20):7219–7235
McFarland KN et al (2014) MeCP2: a novel huntingtin interactor. Hum Mol Genet 23(4):1036–1044
Mingardi J et al (2018) miRNA editing: new insights into the fast control of gene expression in health and disease. Mol Neurobiol 55(10):7717–7727
Mishra J, Chaudhary T, Kumar A (2014) Rosiglitazone synergizes the neuroprotective effects of valproic acid against quinolinic acid-induced neurotoxicity in rats: targeting PPARγ and HDAC pathways. Neurotox Res 26(2):130–151
Mohr AM, Mott JL (2015) Overview of microRNA biology. Semin Liver Dis 35(1):3–11
Moumné L, Betuing S, Caboche J (2013) Multiple aspects of gene dysregulation in Huntington’s disease. Front Neurol 4:127
Nance MA (2017) Genetics of Huntington disease. Handb Clin Neurol 144:3–14
Ng CW et al (2013) Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc Natl Acad Sci U S A 110(6):2354–2359
Packer AN et al (2008) The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci 28(53):14341–14346
Pagiatakis C et al (2021) Epigenetics of aging and disease: a brief overview. Aging Clin Exp Res 33(4):737–745
Pan Y et al (2016) Inhibition of DNA methyltransferases blocks mutant huntingtin-induced neurotoxicity. Sci Rep 6:31022
Parbin S et al (2014) Histone deacetylases: a saga of perturbed acetylation homeostasis in cancer. J Histochem Cytochem 62(1):11–33
Qi X et al (2017) Long non-coding RNA SNHG14 promotes microglia activation by regulating miR-145-5p/PLA2G4A in cerebral infarction. Neuroscience 348:98–106
Ramazi S, Allahverdi A, Zahiri J (2020) Evaluation of post-translational modifications in histone proteins: a review on histone modification defects in developmental and neurological disorders. J Biosci 45:135
Ratovitski T et al (2015) PRMT5-mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington’s disease (HD). Cell Cycle 14(11):1716–1729
Remely M et al (2015) Therapeutic perspectives of epigenetically active nutrients. Br J Pharmacol 172(11):2756–2768
Ryu H et al (2006) ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc Natl Acad Sci U S A 103(50):19176–19181
Saba J et al (2022) Neuroinflammation in Huntington’s disease: a starring role for astrocyte and microglia. Curr Neuropharmacol 20(6):1116–1143
Sadri-Vakili G, Cha JH (2006) Histone deacetylase inhibitors: a novel therapeutic approach to Huntington’s disease (complex mechanism of neuronal death). Curr Alzheimer Res 3(4):403–408
Schaffert LN, Carter WG (2020) Do post-translational modifications influence protein aggregation in neurodegenerative diseases: a systematic review. Brain Sci 10(4):232
Schueller E et al (2020) Dysregulation of histone acetylation pathways in hippocampus and frontal cortex of Alzheimer’s disease patients. Eur Neuropsychopharmacol 33:101–116
Shukla S, Tekwani BL (2020) Histone deacetylases inhibitors in neurodegenerative diseases, neuroprotection and neuronal differentiation. Front Pharmacol 11:537
Siebzehnrübl FA et al (2018) Early postnatal behavioral, cellular, and molecular changes in models of Huntington disease are reversible by HDAC inhibition. Proc Natl Acad Sci U S A 115(37):E8765–E8774
Sinha M, Mukhopadhyay S, Bhattacharyya NP (2012) Mechanism(s) of alteration of micro RNA expressions in Huntington’s disease and their possible contributions to the observed cellular and molecular dysfunctions in the disease. Neuromolecular Med 14(4):221–243
Sixto-López Y, Bello M, Correa-Basurto J (2020) Exploring the inhibitory activity of valproic acid against the HDAC family using an MMGBSA approach. J Comput Aided Mol Des 34(8):857–878
Song L et al (2005) Specific method for the determination of genomic DNA methylation by liquid chromatography-electrospray ionization tandem mass spectrometry. Anal Chem 77(2):504–510
Soragni E et al (2011) Evaluation of histone deacetylase inhibitors as therapeutics for neurodegenerative diseases. Methods Mol Biol 793:495–508
Stack EC et al (2007) Modulation of nucleosome dynamics in Huntington’s disease. Hum Mol Genet 16(10):1164–1175
Subhramanyam CS et al (2019) Microglia-mediated neuroinflammation in neurodegenerative diseases. Semin Cell Dev Biol 94:112–120
Sugama S et al (2009) Possible roles of microglial cells for neurotoxicity in clinical neurodegenerative diseases and experimental animal models. Inflamm Allergy Drug Targets 8(4):277–284
Süssmuth SD et al (2015) An exploratory double-blind, randomized clinical trial with selisistat, a SirT1 inhibitor, in patients with Huntington’s disease. Br J Clin Pharmacol 79(3):465–476
Tafrihi M, Hasheminasab E (2019) MiRNAs: biology, biogenesis, their web-based tools, and databases. Microrna 8(1):4–27
Tan L, Yu JT, Tan L (2015) Causes and consequences of microRNA dysregulation in neurodegenerative diseases. Mol Neurobiol 51(3):1249–1262
Tapias A, Wang ZQ (2017) Lysine acetylation and deacetylation in brain development and neuropathies. Genomics Proteomics Bioinformatics 15(1):19–36
Teleanu DM et al (2022) An overview of oxidative stress, neuroinflammation, and neurodegenerative diseases. Int J Mol Sci 23(11):5938
Thomas EA et al (2008) The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington’s disease transgenic mice. Proc Natl Acad Sci U S A 105(40):15564–15569
Valor LM (2017) Understanding histone deacetylation in Huntington’s disease. Oncotarget 8(4):5660–5661
Valor LM, Guiretti D (2014) What’s wrong with epigenetics in Huntington’s disease? Neuropharmacology 80:103–114
Vashishtha M et al (2013) Targeting H3K4 trimethylation in Huntington disease. Proc Natl Acad Sci U S A 110(32):E3027–E3036
Villar-Menéndez I et al (2013) Increased 5-methylcytosine and decreased 5-hydroxymethylcytosine levels are associated with reduced striatal A2AR levels in Huntington’s disease. Neuromolecular Med 15(2):295–309
Wang F et al (2013) Genome-wide loss of 5-hmC is a novel epigenetic feature of Huntington’s disease. Hum Mol Genet 22(18):3641–3653
Wen Y, Yu Y, Fu X (2017) LncRNA Gm4419 contributes to OGD/R injury of cerebral microglial cells via IκB phosphorylation and NF-κB activation. Biochem Biophys Res Commun 487(4):923–929
Wood H (2013) Neurodegenerative disease: altered DNA methylation and RNA splicing could be key mechanisms in Huntington disease. Nat Rev Neurol 9(3):119
Yang S et al (2020) Lack of RAN-mediated toxicity in Huntington’s disease knock-in mice. Proc Natl Acad Sci U S A 117(8):4411–4417
Yapijakis C (2017) Huntington disease: genetics, prevention, and therapy approaches. Adv Exp Med Biol 987:55–65
Yeh HH et al (2013) Histone deacetylase class II and acetylated core histone immunohistochemistry in human brains with Huntington’s disease. Brain Res 1504:16–24
Ying SY, Chang DC, Lin SL (2018) The microRNA. Methods Mol Biol 1733:1–25
Younesian S et al (2022) The DNA methylation in neurological diseases. Cells 11(21):3439
Zhou Y et al (2016) MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol Neurodegener 11:28
Zimmer-Bensch G (2020) Epigenomic remodeling in Huntington’s disease-master or servant? Epigenomes 4(3):15
Zsindely N, Bodai L (2018) Histone methylation in Huntington’s disease: are bivalent promoters the critical targets? Neural Regen Res 13(7):1191–1192
Zsindely N, Siági F, Bodai L (2021) DNA methylation in Huntington’s disease. Int J Mol Sci 22(23):12736
Zuccato C et al (2003) Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet 35(1):76–83
Funding
The authors disclose that they did not receive any funding, grants, or other forms of support while preparing this manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s inception and design. PT, RA, and PK prepared the materials, collected data, and analysed the results. PT and RA wrote the first draught of the manuscript, and PK provided feedback on prior draughts. The final manuscript was read and approved by all authors.
Corresponding author
Ethics declarations
Conflict of interest
There are no relevant financial or non-financial interests to disclose for the authors.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Temgire, P., Arthur, R. & Kumar, P. Neuroinflammation and the role of epigenetic-based therapies for Huntington’s disease management: the new paradigm. Inflammopharmacol (2024). https://doi.org/10.1007/s10787-024-01477-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10787-024-01477-0