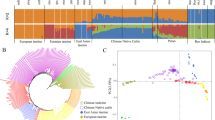
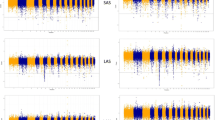
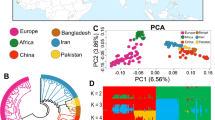
Abstract
Improving the low productivity levels of native cattle breeds in smallholder farming systems is a pressing concern in Pakistan. Crossbreeding high milk-yielding holstein friesian (HF) breed with the adaptability and heat tolerance of Sahiwal cattle has resulted in offspring that are well-suited to local conditions and exhibit improved milk yield. The exploration of how desirable traits in crossbred dairy cattle are selected has not yet been investigated. This study aims to provide the first overview of the selective pressures on the genome of crossbred dairy cattle in Pakistan. A total of eighty-one crossbred, thirty-two HF and twenty-four Sahiwal cattle were genotyped, and additional SNP genotype data for HF and Sahiwal were collected from a public database to equate the sample size in each group. Within-breed selection signatures in crossbreds were investigated using the integrated haplotype score. Crossbreds were also compared to each of their parental breeds to discover between-population signatures of selection using two approaches: cross-population extended haplotype homozygosity and fixation index. We identified several overlapping genes associated with production, immunity, and adaptation traits, including U6, TMEM41B, B4GALT7, 5S_rRNA, RBM27, POU4F3, NSD1, PRELID1, RGS14, SLC34A1, TMED9, B4GALT7, OR2AK3, OR2T16, OR2T60, OR2L3, and CTNNA1. Our results suggest that regions responsible for milk traits have generally experienced stronger selective pressure than others.








Similar content being viewed by others
Data Availability
All relevant data are within the paper and its supplementary data files.
References
Abo-Ismail MK, Vander Voort G, Squires JJ, Swanson KC, Mandell IB, Liao X, Stothard P, Moore S, Plastow G, Miller SP (2014) Single nucleotide polymorphisms for feed efficiency and performance in crossbred beef cattle. BMC Genet 15(1):14
Ahmad SF, Panigrahi M, Chhotaray S, Pal D, Chauhan A, Sonwane A, Parida S, Bhushan B, Gaur GK, Mishra BP (2020) Population structure and admixture analysis in frieswal crossbred cattle of India—a pilot study. Anim Biotechnol 31:86–92
Akey JM, Zhang G, Zhang K, Jin L, Shriver MD (2002) Interrogating a high-density SNP map for signatures of natural selection. Genome Res 12:1805–1814
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664
Aliloo H, Mrode R, Okeyo A, Gibs ONJ (2019) Signatures of selection in admixed dairy cattle of Kenya. Proceedings of the 23rd Conference of the Association for the Advancement of Animal Breeding and Genetics (AAABG), Armidale, New South Wales, Australia, 27th October-1st November 2019. Association for the Advancement of Animal Breeding and Genetics, 262–265
Alvarenga AB, Oliveira HR, Chen S-Y, Miller SP, Marchant-Forde JN, Grigoletto L, Brito LF (2021) A systematic review of genomic regions and candidate genes underlying behavioral traits in farmed mammals and their link with human disorders. Animals 11:715
Álvarez Cecco P, Rogberg Munoz A, Balbi M, Bonamy M, Munilla S, Forneris NS, Peral Garcia P, Cantet RJ, Giovambattista G, Fernández ME (2022) Genome-wide scan for signatures of selection in the Brangus cattle genome. J Anim Breed Genet 139(679):694
Andersson L, Georges M (2004) Domestic-animal genomics: deciphering the genetics of complex traits. Nat Rev Genet 5:202–212
Anonymous (2023) Economic Survey of Pakistan, 2022–2023, Chapter 22, Ministry of Finance, Government of Pakistan, pp 19–39
Ariel O, Gendron D, Dudemaine P-L, Gévry N, Ibeagha-Awemu EM, Bissonnette N (2020) Transcriptome profiling of bovine macrophages infected by Mycobacterium avium spp. paratuberculosis depicts foam cell and innate immune tolerance phenotypes. Front Immunol. https://doi.org/10.3389/fimmu.2019.02874
Asselstine V, Medrano J, Cánovas A (2022) Identification of novel alternative splicing associated with mastitis disease in Holstein dairy cows using large gap read mapping. BMC Genom 23:1–15
Bahbahani H, Afana A, Wragg D (2018) Genomic signatures of adaptive introgression and environmental adaptation in the Sheko cattle of southwest Ethiopia. PLoS ONE 13:e0202479
Bang NN, Hayes BJ, Lyons RE, Randhawa IA, Gaughan JB, McNeill DM (2022) Genomic diversity and breed composition of Vietnamese smallholder dairy cows. J Anim Breed Genet 139:145–160
Barendse W, Harrison BE, Bunch RJ, Thomas MB, Turner LB (2009) Genome wide signatures of positive selection: the comparison of independent samples and the identification of regions associated to traits. BMC Genom 10:1–15
Bebe BO, Udo HM, Rowlands GJ, Thorpe W (2003) Smallholder dairy systems in the Kenya highlands: breed preferences and breeding practices. Livest Prod Sci 82:117–127
Bennewitz J, Reinsch N, Grohs C, Levéziel H, Malafosse A, Thomsen H, Xu N, Looft C, Kühn C, Brockmann GA (2003) Combined analysis of data from two granddaughter designs: a simple strategy for QTL confirmation and increasing experimental power in dairy cattle. Genet Sel Evol 35:1–20
Bhatia G, Patterson N, Sankararaman S, Price AL (2013) Estimating and interpreting FST: the impact of rare variants. Genome Res 23:1514–1521
Bickhart DM, Liu GE (2013) Identification of candidate transcription factor binding sites in the cattle genome. Genom Proteom Bioinform 11:195–198
Browning BL, Zhou Y, Browning SR (2018) A one-penny imputed genome from next-generation reference panels. American J Hum Genet 103:338–348
Buitenhuis B, Janss LL, Poulsen NA, Larsen LB, Larsen MK, Sørensen P (2014) Genome-wide association and biological pathway analysis for milk-fat composition in Danish Holstein and Danish Jersey cattle. BMC Genom 15:1–11
Buzanskas ME, Grossi DDA, Ventura RV, Schenkel FS, Chud TCS, Stafuzza NB, Rola LD, Meirelles SLC, Mokry FB, Mudadu MDA (2017) Candidate genes for male and female reproductive traits in Canchim beef cattle. J Anim Sci Biotechnol 8:1–10
Cai Z, Guldbrandtsen B, Lund MS, Sahana G (2019) Dissecting closely linked association signals in combination with the mammalian phenotype database can identify candidate genes in dairy cattle. BMC Genet 20:1–12
Carvalho M, Baldi F, Alexandre PA, Santana MHDA, Ventura RV, Bueno RS, Bonin MDN, Rezende FMD, Coutinho LL, Eler JP (2019) Genomic regions and genes associated with carcass quality in Nelore cattle. Genet Mol Res 18:1–15
Cheruiyot EK, Bett RC, Amimo JO, Zhang Y, Mrode R, Mujibi FD (2018) Signatures of selection in admixed dairy cattle in Tanzania. Front Genet 9:607
Chhotaray S, Panigrahi M, Pal D, Ahmad SF, Bhanuprakash V, Kumar H, Parida S, Bhushan B, Gaur G, Mishra B (2021) Genome-wide estimation of inbreeding coefficient, effective population size and haplotype blocks in Vrindavani crossbred cattle strain of India. Biol Rhythm Res 52:666–679
Ciganda M, Prohaska K, Hellman K, Williams N (2012) A novel association between two trypanosome-specific factors and the conserved L5–5S rRNA complex. PLoS ONE. https://doi.org/10.1371/journal.pone.0041398
Cochran SD, Cole JB, Null DJ, Hansen PJ (2013) Discovery of single nucleotide polymorphisms in candidate genes associated with fertility and production traits in Holstein cattle. BMC Genet 14:1–23
Cole JB, Wiggans GR, Ma L, Sonstegard TS, Lawlor TJ, Crooker BA, van Tassell CP, Yang J, Wang S, Matukumalli LK (2011) Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary US Holstein cows. BMC Genomics 12:1–17
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, Depristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158
Dayo GK, Thevenon S, Berthier D, Moazami-Goudarzi K, Denis C, Cuny G, Eggen A, Gautier M (2009) Detection of selection signatures within candidate regions underlying trypanotolerance in outbred cattle populations. Mol Ecol 18:1801–1813
Delosière M, Pires J, Bernard L, Cassar-Malek I, Bonnet M (2019) Milk proteome from in silico data aggregation allows the identification of putative biomarkers of negative energy balance in dairy cows. Sci Rep 9:1–11
Devani K, Crowley JJ, Plastow G, Orsel K, Valente TS (2021) Genetic parameter estimations and genomic insights for teat and udder structure in young and mature Canadian angus cows. J Anim Sci. https://doi.org/10.1093/jas/skab087
Do D, Schenkel F, Miglior F, Zhao X, Ibeagha-Awemu E (2020) Targeted genotyping to identify potential functional variants associated with cholesterol content in bovine milk. Anim Genet 51:200–209
Edea Z, Jeoung YH, Shin S-S, Ku J, Seo S, Kim I-H, Kim S-W, Kim K-S (2018) Genome–wide association study of carcass weight in commercial Hanwoo cattle. Asian Australas J Anim Sci 31:327
Elavarasan K, Kumar S, Agarwal S, Vani A, Sharma R, Kumar S, Chauhan A, Sahoo NR, Verma MR, Gaur GK (2023) Estimation of microsatellite-based autozygosity and its correlation with pedigree inbreeding coefficient in crossbred cattle. Anim Biotechnol 34(8):3564–3577
FAO (2011) Mapping supply and demand for animal-source foods to 2030. Animal Production and Health Working Paper 2: 1–54
Galliou JM, Kiser JN, Oliver KF, Seabury CM, Moraes JG, Burns GW, Spencer TE, Dalton J, Neibergs HL (2020) Identification of loci and pathways associated with heifer conception rate in US Holsteins. Genes 11:767
Gao Y, Jiang J, Yang S, Hou Y, Liu GE, Zhang S, Zhang Q, Sun D (2017) CNV discovery for milk composition traits in dairy cattle using whole genome resequencing. BMC Genom 18:1–12
Gautier M, Naves M (2011) Footprints of selection in the ancestral admixture of a new world Creole cattle breed. Mol Ecol 20:3128–3143
Gautier M, Klassmann A, Vitalis R (2017) rehh 2.0: a reimplementation of the R package rehh to detect positive selection from haplotype structure. Mol Ecol Resour 17:78–90
Ghoreishifar SM, Eriksson S, Johansson AM, Khansefid M, Moghaddaszadeh-Ahrabi S, Parna N, Davoudi P, Javanmard A (2020a) Signatures of selection reveal candidate genes involved in economic traits and cold acclimation in five Swedish cattle breeds. Genet Sel Evol 52:1–15
Ghoreishifar SM, Moradi-Shahrbabak H, Fallahi MH, Jalil Sarghale A, Moradi-Shahrbabak M, Abdollahi-Arpanahi R, Khansefid M (2020b) Genomic measures of inbreeding coefficients and genome-wide scan for runs of homozygosity islands in Iranian river buffalo. Bubalus Bubalis BMC Genet 21(1):12
Hayes BJ, Daetwyler HD (2019) 1000 Bull genomes project to map simple and complex genetic traits in cattle: applications and outcomes. Annu Rev Anim Biosci 7:89–102
de Heras-SaldanaLas S, Lopez BI, Moghaddar N, Park W, Park JE, Chung KY, Lim D, Lee SH, Shin D, van Der Werf JH (2020) Use of gene expression and whole-genome sequence information to improve the accuracy of genomic prediction for carcass traits in Hanwoo cattle. Genet Sel Evol 52(1):16
Hou Y, Liu GE, Bickhart DM, Cardone MF, Wang K, Kim E-S, Matukumalli LK, Ventura M, Song J, Vanraden PM (2011) Genomic characteristics of cattle copy number variations. BMC Genom 12:1–11
Hu Y, Xia H, Li M, Xu C, Ye X, Su R, Zhang M, Nash O, Sonstegard TS, Yang L (2020) Comparative analyses of copy number variations between Bos taurus and Bos indicus. BMC Genomics 21:1–11
Huang W, Peñagaricano F, Ahmad K, Lucey J, Weigel K, Khatib H (2012) Association between milk protein gene variants and protein composition traits in dairy cattle. J Dairy Sci 95:440–449
Huang Y-Z, Wang Q, Zhang C-L, Fang X-T, Song E-L, Chen H (2016) Genetic variants in SDC3 gene are significantly associated with growth traits in two Chinese beef cattle breeds. Anim Biotechnol 27:190–198
Iqbal N, Liu X, Yang T, Huang Z, Hanif Q, Asif M, Khan QM, Mansoor S (2019) Genomic variants identified from whole-genome resequencing of indicine cattle breeds from Pakistan. PLoS ONE 14:e0215065
Jiang J, Ma L, Prakapenka D, Vanraden PM, Cole JB, Da Y (2019) A large-scale genome-wide association study in US Holstein cattle. Front Genet. https://doi.org/10.3389/fgene.2019.00412
Júnior GO, Perez B, Cole J, Santana MHDA, Silveira J, Mazzoni G, Ventura RV, Júnior MS, Kadarmideen HN, Garrick DJ (2017) Genomic study and medical subject headings enrichment analysis of early pregnancy rate and antral follicle numbers in Nelore heifers. J Anim Sci 95:4796–4812
Kang H, Zhao D, Xiang H, Li J, Zhao G, Li H (2021) Large-scale transcriptome sequencing in broiler chickens to identify candidate genes for breast muscle weight and intramuscular fat content. Genet Sel Evol 53:1–23
Khayatzadeh N, Mészáros G, Utsunomiya Y, Garcia J, Schnyder U, Gredler B, Curik I, Sölkner J (2016) Locus-specific ancestry to detect recent response to selection in admixed Swiss Fleckvieh cattle. Anim Genet 47:637–646
Kolbehdari D, Wang Z, Grant J, Murdoch B, Prasad A, Xiu Z, Marques E, Stothard P, Moore S (2009) A whole genome scan to map QTL for milk production traits and somatic cell score in Canadian Holstein bulls. J Anim Breed Genet 126:216–227
Kumar S, Alex R, Gaur G, Mukherjee S, Mandal D, Singh U, Tyagi S, Kumar A, Das A, Deb R (2018) Evolution of Frieswal cattle: a crossbred dairy animal of India. Indian J Anim Sci 88:265–275
Kwon T, Kim K, Caetano-Anolles K, Sung S, Cho S, Jeong C, Hanotte O, Kim H (2022) Mitonuclear incompatibility as a hidden driver behind the genome ancestry of African admixed cattle. BMC Biol 20:20
Lan X, Peñagaricano F, Dejung L, Weigel K, Khatib H (2013) A missense mutation in the PROP1 (prophet of Pit 1) gene affects male fertility and milk production traits in the US Holstein population. J Dairy Sci 96:1255–1257
Leal-Gutiérrez JD, Elzo MA, Johnson DD, Hamblen H, Mateescu RG (2019) Genome wide association and gene enrichment analysis reveal membrane anchoring and structural proteins associated with meat quality in beef. BMC Genom 20:1–18
Lee SH, Choi BH, Lim D, Gondro C, Cho YM, Dang CG, Sharma A, Jang GW, Lee KT, Yoon D (2013) Genome-wide association study identifies major loci for carcass weight on BTA14 in Hanwoo (Korean cattle). PLoS ONE 8:e74677
Lee H-J, Chung Y, Chung KY, Kim Y-K, Lee JH, Koh YJ, Lee SH (2022) Use of a graph neural network to the weighted gene co-expression network analysis of Korean native cattle. Sci Rep 12:9854
Leroy G, Baumung R, Boettcher P, Scherf B, Hoffmann I (2016) Sustainability of crossbreeding in developing countries; definitely not like crossing a meado. Animal 10:262–273
Liu D, Chen Z, Zhao W, Guo L, Sun H, Zhu K, Liu G, Shen X, Zhao X, Wang Q (2021) Genome-wide selection signatures detection in Shanghai Holstein cattle population identified genes related to adaption, health and reproduction traits. BMC Genom 22:1–19
Magee DA, Sikora KM, Berkowicz EW, Berry DP, Howard DJ, Mullen MP, Evans RD, Spillane C, Machugh DE (2010) DNA sequence polymorphisms in a panel of eight candidate bovine imprinted genes and their association with performance traits in Irish Holstein-Friesian cattle. BMC Genet 11:1–15
Makoni N, Mwai R, Redda T, van der Zijpp A, van der Lee J (2014) White gold: Opportunities for dairy sector development collaboration in East Africa. Centre for Development Innovation, Wageningen
Marshall K, Gibson JP, Mwai O, Mwacharo JM, Haile A, Getachew T, Mrode R, Kemp SJ (2019) Livestock genomics for developing countries—African examples in practice. Front Genet 10:297
Mbole-Kariuki MN, Sonstegard T, Orth A, Thumbi S, Bronsvoort BDC, Kiara H, Toye P, Conradie I, Jennings A, Coetzer K (2014) Genome-wide analysis reveals the ancient and recent admixture history of East African Shorthorn Zebu from Western Kenya. Heredity 113:297–305
Moretti R, Soglia D, Chessa S, Sartore S, Finocchiaro R, Rasero R, Sacchi P (2021) Identification of SNPs associated with somatic cell score in candidate genes in Italian Holstein Friesian bulls. Animals 11:366
Mumtaz S, Mukherjee A, Pathak P, Parveen K (2021) Effects of inbreeding on performance traits in Karan Fries crossbred cattle. Indian J Anim Sci. https://doi.org/10.56093/ijans.v91i5.115396
Naserkheil M, Bahrami A, Lee D, Mehrban H (2020) Integrating single-step GWAS and bipartite networks reconstruction provides novel insights into yearling weight and carcass traits in Hanwoo beef cattle. Animals 10:1836
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Rocha D, Billerey C, Samson F, Boichard D, Boussaha M (2014) Identification of the putative ancestral allele of bovine single-nucleotide polymorphisms. J Anim Breed Genet 131:483–486
Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R (2007) Genome-wide detection and characterization of positive selection in human populations. Nature 449:913–918
Sanchez M-P, Rocha D, Charles M, Boussaha M, Hozé C, Brochard M, Delacroix-Buchet A, Grosperrin P, Boichard D (2021) Sequence-based GWAS and post-GWAS analyses reveal a key role of SLC37A1, ANKH, and regulatory regions on bovine milk mineral content. Sci Rep 11:1–15
Saravanan K, Panigrahi M, Kumar H, Parida S, Bhushan B, Gaur G, Kumar P, Dutt T, Mishra B, Singh R (2022) Genome-wide assessment of genetic diversity, linkage disequilibrium and haplotype block structure in Tharparkar cattle breed of India. Anim Biotechnol 33:297–311
Seo M, Caetano-Anolles K, Rodriguez-Zas S, Ka S, Jeong JY, Park S, Kim MJ, Nho W-G, Cho S, Kim H (2016) Comprehensive identification of sexually dimorphic genes in diverse cattle tissues using RNA-seq. BMC Genom 17:1–18
Singh A, Mehrotra A, Gondro C, Romero ARDS, Pandey AK, Karthikeyan A, Bashir A, Mishra B, Dutt T, Kumar A (2020) Signatures of selection in composite Vrindavani cattle of India. Front Genet 11:589496
Singh A, Kumar A, Gondro C, da Silva Romero AR, Karthikeyan A, Mehrotra A, Pandey AK, Dutt T, Mishra BP (2021) Identification of genes affecting milk fat and fatty acid composition in Vrindavani crossbred cattle using 50 K SNP-Chip. Trop Ani Health Prod 53:1–8
Team RC (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
THORNTON, P. K. (2010) Livestock production: recent trends, future prospects. Philos Transact Royal Soc B: Biol Sci 365:2853–2867
Vanvanhossou SFU, Scheper C, Dossa LH, Yin T, Brügemann K, König S (2020) A multi-breed GWAS for morphometric traits in four Beninese indigenous cattle breeds reveals loci associated with conformation, carcass and adaptive traits. BMC Genom 21:1–16
Voight BF, Kudaravalli S, Wen X, Pritchard JK (2006) A map of recent positive selection in the human genome. PLoS Biol 4:e72
Ward JA, McHugo GP, Dover MJ, Hall TJ, Ismael Ng’ang’a S, Sonstegard TS, Bradley DG, Frantz LA, Salter-Townshend M, MacHugh DE (2022) Genome-wide local ancestry and evidence for mitonuclear coadaptation in African hybrid cattle populations. Iscience 25(7):104672
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution. https://doi.org/10.2307/2408641
Xerxa E, Barbisin M, Chieppa MN, Krmac H, Vallino Costassa E, Vatta P, Simmons M, Caramelli M, Casalone C, Corona C (2016) Whole blood gene expression profiling in preclinical and clinical cattle infected with atypical bovine spongiform encephalopathy. PLoS ONE 11:e0153425
Xu Y, Han Q, Ma C, Wang Y, Zhang P, Li C, Cheng X, Xu H (2021) Comparative proteomics and phosphoproteomics analysis reveal the possible breed difference in yorkshire and duroc boar spermatozoa. Front Cell Dev Biol 9:652809
Yang B, Wang J, Wang Y, Zhou H, Wu X, Tian Z, Sun B (2013) Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J Immunol 190:3613–3619
Yuan Z, Chen Y, Chen Q, Guo M, Kang L, Zhu G, Jiang Y (2016) Characterization of chicken MMP13 expression and genetic effect on egg production traits of its promoter polymorphisms. G3: Genes Genom Genet 6:1305–1312
Zhang Q, Calus MP, Bosse M, Sahana G, Lund MS, Guldbrandtsen B (2018) Human-mediated introgression of haplotypes in a modern dairy cattle breed. Genetics 209:1305–1317
Zhang Y, Gargan S, Lu Y, Stevenson NJ (2021) An overview of current knowledge of deadly CoVs and their interface with innate immunity. Viruses 13:560
Zhang S, Yao Z, Li X, Zhang Z, Liu X, Yang P, Chen N, Xia X, Lyu S, Shi Q (2022) Assessing genomic diversity and signatures of selection in Pinan cattle using whole-genome sequencing data. BMC Genom 23:460
Acknowledgements
The authors express their gratitude to Army Farm, Renalakhurd, for their assistance during sample collection, and Dr. Akansha Singh from India, for her assistance during analysis. Additionally, they acknowledge the Higher Education Commission (HEC) for providing funding to the first author’s PhD studies
Funding
The current study was supported by Pakistan Agricultural Research Council, Agricultural Linkages Programme (ALP), Project#AS 016 and initiated by National Institute for Biotechnology and Genetic Engineering (NIBGE).
Author information
Authors and Affiliations
Contributions
Conceptualization: Fakhar un Nisa, Raphael Mrode. Funding acquisition: Shahid Mansoor, Muhammad Asif, Imran Amin. Data curation: Fakhar Un Nisa, Muhammad Asif, Fazeela Arshad. Methodology: Fakhar un Nisa, Iram Ilyas. Formal Analysis: Fakhar un Nisa, Rubab Zahra Naqvi. Project administration: Shahid Mansoor, Raphael Mrode, Zahid Mukhtar. Manuscript Initial drafting: Fakhar un Nisa, Manuscript review & editing: Zahid Mukhtar, Raphael Mrode, Shahid Mansoor, Imran Amin.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that there is no conflict of interest as defined by Springer or other interests that might be perceived to influence the results/or discussion reported in this paper.
Ethical Approval
The ethical review and approval of the animal study was conducted by the Institutional Animal Ethics Committee at the National Institute for Biotechnology and Genetic Engineering College, PIEAS, Faisalabad Pakistan.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nisa, F.u., Naqvi, R.Z., Arshad, F. et al. Assessment of Genomic Diversity and Selective Pressures in Crossbred Dairy Cattle of Pakistan. Biochem Genet (2024). https://doi.org/10.1007/s10528-024-10809-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10528-024-10809-2