Abstract
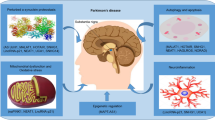
Parkinson’s disease (PD) is known as one of the most common degenerative disorders related to the damage of the central nervous system (CNS). This brain disorder is also characterized by the formation of Lewy bodies in the cytoplasm of the dopaminergic neurons in the substantia nigra pars compacta (SNc), which consequently leads to motor and non-motor symptoms. With regard to the growing trend in the number of cases with PD and its effects on individuals, families, and communities, immediate treatments together with diagnostic methods are required. In this respect, long non-coding ribonucleic acids (lncRNAs) represent a large class of ncRNAs with more than 200 nucleotides in length, playing key roles in some important processes including gene expression, cell differentiation, genomic imprinting, apoptosis, and cell cycle. They are highly expressed in the CNS and previous studies have further reported that the expression profile of lncRNAs is disrupted in human diseases such as neurodegenerative disorders. Since the levels of some lncRNAs change over time in the brains of patients with PD, a number of previous studies have examined their potentials as biomarkers for this brain disorder. Therefore, the main purpose of this study was to review the advances in the related literature on lncRNAs as diagnostic, therapeutic, and prognostic biomarkers for PD.

adapted from Ying Lyu et al. (2019). The functions of lncRNAs involved in the regulation of several processes at different levels, including (1) transcription, (2) post-transcription, and (3) interaction with other biological molecules
Similar content being viewed by others
Abbreviations
- PD:
-
Parkinson’s disease
- CNS:
-
Central nervous system
- SNc:
-
Substantia nigra pars compacta
- SNCA:
-
Alpha-synuclein
- LRRK2:
-
Leucine-rich repeat kinase 2
- RNA:
-
Ribonucleic acid
- lncRNA:
-
Long non-coding RNA
- HD:
-
Huntington’s disease
- AD:
-
Alzheimer’s disease
- NEAT1:
-
Nuclear enriched assembly transcript 1
- SNHG1:
-
Small nucleolar RNA host gene 1
- HTR2A:
-
5-Hydroxytryptamine receptor 2A
- NOD:
-
Nucleotide-binding domain
- UCA1:
-
Urothelial carcinoma-associated 1
- MALAT1:
-
Metastasis-associated lung adenocarcinoma transcript 1
- lincRNA-p21:
-
Long intergenic non-coding RNA p21
- HIF1α:
-
Hypoxia-inducible factor 1 subunit alpha
- linc-POU3F3:
-
Long intergenic non-coding RNA POU3F3
- L1CAM:
-
L1 cell adhesion molecule
- GCase:
-
Glucocerebrosidase
- miR-15a-5p:
-
MicroRNA-15a-5p
- C57BL/6:
-
C57 black 6
- MPTP:
-
1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine
- CASP3:
-
Caspase-3
- NGF:
-
Neuronal growth factor
- BDNF:
-
Brain-derived neurotrophic factor
- IL-1β:
-
Interleukin 1 beta
- TNF-α:
-
Tumor necrosis factor alpha
- PI3K:
-
Phosphatidylinositol 3-kinase;
- AKT:
-
Protein kinase B
- IκBα:
-
Inhibitor of nuclear factor kappa B
- UCHL1:
-
Ubiquitin carboxy-terminal hydrolase L1
- NURR1:
-
Nuclear receptor-related 1 protein
- AS:
-
Antisense
- BACE1:
-
β-Site amyloid precursor protein cleaving enzyme 1
- iNOS:
-
Inducible nitric oxide synthase
- MAP1LC3:
-
Microtubule-associated protein 1 light chain 3
- PINK1:
-
PTEN-induced kinase 1
- ATP:
-
Adenosine triphosphate
- ROS:
-
Reactive oxygen species
- LDH:
-
Lactate dehydrogenase
- SOD:
-
Superoxide dismutase
- RNAi:
-
RNA interference
- ASO:
-
AS oligonucleotide
- HOTAIR:
-
HOX transcript AS intergenic RNA
- NLRP3:
-
NOD-like receptor protein 3
- N2a:
-
Neuro 2A
- NRF2:
-
Nuclear factor (erythroid-derived 2)-like-2 factor
- EZH2:
-
Enhancer of zeste homolog 2
- PRC2:
-
Polycomb repressive complex 2
- LRRK2:
-
Leucine-rich repeat kinase
References
Pringsheim T, Jette N, Frolkis A, Steeves T (2014) The prevalence of Parkinson’s disease: a systematic review and meta-analysis
Hauser DN, Hastings TG (2013) Mitochondrial dysfunction and oxidative stress in Parkinson’s disease and monogenic parkinsonism. Neurobiol Dis 51:35
Bonifati V (2014) Genetics of Parkinson’s disease–state of the art, 2013. Parkinsonism Relat Disord 20:S23
Renani P, Taheri F, Rostami D, Farahani N, Abdolkarimi H, Abdollahi E et al (2019) Involvement of aberrant regulation of epigenetic mechanisms in the pathogenesis of Parkinson’s disease and epigenetic-based therapies. J Cell Physiol 234(11):19307
Bradaric BD, Patel A, Schneider JA, Carvey PM, Hendey B (2012) Evidence for angiogenesis in Parkinson’s disease, incidental Lewy body disease, and progressive supranuclear palsy. J Neural Transm 119(1):59
Jankovic J (2008) Parkinson’s disease: clinical features and diagnosis. J Neurol Neurosurg Psychiatry 79(4):368
Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y et al (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47(3):199
Kopp F, Mendell JT (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172(3):393–407
Quinn J, Chang H (2016) Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17(1):47
Managadze D, Lobkovsky AE, Wolf YI, Shabalina SA, Rogozin IB, Koonin EV (2013) The vast, conserved mammalian lincRNome. PLoS Comput Bio. 9(2).
Qureshi IA, Mehler MF (2013) Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics 10(4):632–646
Kadakkuzha BM, Liu X-A, McCrate J, Shankar G, Rizzo V, Afinogenova A, et al. (2015) Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front Cell Neurosci; 9
Ponjavic J, Oliver PL, Lunter G, Ponting CP (2009) Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet;5(8):e1000617-e.
Zhou Y, Gu C, Li J, Zhu L, Huang G, Dai J et al (2018) Aberrantly expressed long noncoding RNAs and genes in Parkinson’s disease. Neuropsychiatr Dis Treat 14:3219
Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat rev Neurosci 13(8):528
Schapira AH (2013) Recent developments in biomarkers in Parkinson disease. Curr Opin Neurol 26(4):395
Schapira AH, Tolosa E (2010) Molecular and clinical prodrome of Parkinson disease: implications for treatment. Nat Rev Neurol 6(6):309–317
Bonanni L, Thomas A, Onofrj M (2006) Diagnosis and management of dementia with Lewy bodies: third report of the DLB Consortium. Neurology 66(9):1455 (author reply)
Kraus T, Haider M, Spanner J, Steinmaurer M, Dietinger V, Kretzschmar H (2017) Altered long noncoding RNA expression precedes the course of Parkinson’s disease-a preliminary report. Mol Neurobiol 54(4):2869
Soreq L, Guffanti A, Salomonis N, Simchovitz A, Israel Z, Bergman H, et al. (2014) Long non-coding RNA and alternative splicing modulations in Parkinson’s leukocytes identified by RNA sequencing. PLoS Comput Bio 10(3)
Lv Q, Wang Z, Zhong Z, Huang W (2020) Role of long noncoding RNAs in Parkinson’s disease: putative biomarkers and therapeutic targets. Parkinson’s Disease 2020
You J, Fang N, Gu J, Zhang Y, Li X, Zu L et al (2014) Noncoding RNA small nucleolar RNA host gene 1 promote cell proliferation in nonsmall cell lung cancer. Indian Journal cancer. 51:e99
Hall J, Messenger Z, Tam H, Phillips S, Recio L, Smart R (2015) Long noncoding RNA lincRNA-p21 is the major mediator of UVB-induced and p53-dependent apoptosis in keratinocytes. Cell Death & Dis. 6(3):e1700
Zou J, Guo Y, Wei L, Yu F, Yu B, Xu A (2020) Long noncoding RNA POU3F3 and α-synuclein in plasma L1CAM exosomes combined with β-glucocerebrosidase activity: potential predictors of Parkinson’s disease. Neurotherapeutics: the journal of the American Society for Experimental NeuroTherapeutics
Shi M, Liu C, Cook TJ, Bullock KM, Zhao Y, Ginghina C et al (2014) Plasma exosomal α-synuclein is likely CNS-derived and increased in Parkinson’s disease. Acta Neuropathol 128(5):639
Lv Q, Wang Z, Zhong Z, Huang W (2020) Role of long noncoding RNAs in Parkinson’s disease: putative biomarkers and therapeutic targets. Parkinson’s Disease 2020
Jamebozorgi K, Taghizadeh E, Rostami D, Pormasoumi H, Barreto G, Hayat S et al (2019) Cellular and molecular aspects of Parkinson treatment: future therapeutic perspectives. Mol Neurobiol 56(7):4799
Fonseca-Ornelas L, Eisbach S, Paulat M, Giller K, Fernández C, Outeiro T et al (2014) Small molecule-mediated stabilization of vesicle-associated helical α-synuclein inhibits pathogenic misfolding and aggregation. Nat Commun 5:5857
Chen Y, Lian Y, Ma Y, Wu C, Zheng Y, Xie N (2018) LncRNA SNHG1 promotes α-synuclein aggregation and toxicity by targeting miR-15b-5p to activate SIAH1 in human neuroblastoma SH-SY5Y cells. Neurotoxicology 68:212
Chaturvedi RK, Flint BM (2013) Mitochondrial diseases of the brain. Free Radical Biol Med 63:1–29
Ni Y, Huang H, Chen Y, Cao M, Zhou H, Zhang Y (2017) Investigation of long non-coding RNA expression profiles in the substantia nigra of Parkinson’s disease. Cell Mol Neurobiol 37(2):329–338
Ni Y, Huang H, Chen Y, Cao M, Zhou H, Zhang Y (2017) Investigation of long non-coding RNA expression profiles in the substantia nigra of Parkinson’s disease. Cell Mol Neurobiol 37(2):329
Wen J, Ma Y, Yang G, Wang G (2017) Analysis of circulating long non-coding RNA UCA1 as potential biomarkers for diagnosis and prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci 21(3):498
Wang Y, Chen W, Yang C, Wu W, Wu S, Qin X et al (2012) Long non-coding RNA UCA1a (CUDR) promotes proliferation and tumorigenesis of bladder cancer. Int J Oncol 41(1):276
Lu M, Sun W, Shen J, Wei M, Chen B, Qi Y et al (2018) LncRNA-UCA1 promotes PD development by upregulating SNCA. Eur Rev Med Pharmacol Sci 22(22):7908
Cai L, Tu L, Li T, Yang X, Ren Y, Gu R et al (2019) Downregulation of lncRNA UCA1 ameliorates the damage of dopaminergic neurons, reduces oxidative stress and inflammation in Parkinson’s disease through the inhibition of the PI3K/Akt signaling pathway. Int immunopharmacology. 75:105734
Liu Z, Meray RK, Grammatopoulos TN, Fredenburg RA, Cookson MR, Liu Y et al (2009) Membrane-associated farnesylated UCH-L1 promotes α-synuclein neurotoxicity and is a therapeutic target for Parkinson’s disease. Proc Natl Acad Sci USA 106(12):4635
Grimes D, Han F, Panisset M, Racacho L, Xiao F, Zou R et al (2006) Translated mutation in the Nurr1 gene as a cause for Parkinson’s disease. Mov dis: offic J Mov Dis Soc 21(7):906
Gong B, Cao Z, Zheng P, Vitolo OV, Liu S, Staniszewski A et al (2006) Ubiquitin hydrolase Uch-L1 rescues β-amyloid-induced decreases in synaptic function and contextual memory. Cell 126(4):775–788
Gong B, Cao Z, Zheng P, Vitolo O, Liu S, Staniszewski A et al (2006) Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell 126(4):775
Carrieri C, Forrest AR, Santoro C, Persichetti F, Carninci P, Zucchelli S, et al (2015) Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front Cell Neurosci 9
Riva P, Ratti A, Venturin M (2016) The long non-coding RNAs in neurodegenerative diseases: novel mechanisms of pathogenesis. Curr Alzheimer Res 13(11):1219
Li Y, Fang J, Zhou Z, Zhou Q, Sun S, Jin Z, et al (2020) Downregulation of lncRNA BACE1-AS improves dopamine-dependent oxidative stress in rats with Parkinson’s disease by upregulating microRNA-34b-5p and downregulating BACE1. Cell Cycle (Georgetown, Tex) 1–14
Scrivo A, Bourdenx M, Pampliega O, Cuervo AM (2018) Selective autophagy as a potential therapeutic target for neurodegenerative disorders. Lancet Neurol 17(9):802
Tanida I, Ueno T, Kominami E (2008) LC3 and Autophagy. Methods in molecular biology (Clifton, NJ) 445:77
Xie S, Zhou F, Li J, Duan S (2019) NEAT1 regulates MPP+-induced neuronal injury by targeting miR-124 in neuroblastoma cells. Neurosci lett. 708:134340
Simchovitz A, Hanan M, Niederhoffer N, Madrer N, Yayon N, Bennett ER et al (2019) NEAT1 is overexpressed in Parkinson’s disease substantia nigra and confers drug-inducible neuroprotection from oxidative stress. FASEB J 33(10):11223–11234
Yan W, Chen Z, Chen J, Chen H (2018) LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson’s disease through stabilizing PINK1 protein. Biochem Biophys Res Commun 496(4):1019
Kawajiri S, Saiki S, Sato S, Sato F, Hatano T, Eguchi H et al (2010) PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett 584(6):1073
Li L, Xu J, Wu M, Hu J (2018) Protective role of microRNA-221 in Parkinson’s disease. Bratisl Lek Listy 119(1):22
Geng L, Zhao J, Liu W, Chen Y (2019) Knockdown of NEAT1 ameliorated MPP+-induced neuronal damage by sponging miR-221 in SH-SY5Y cells. RSC Adv 9(43):25257–25265
Xu X, Cui L, Zhong W, Cai Y (2020) Autophagy-associated lncRNAs: promising targets for neurological disease diagnosis and therapy. Neural Plast
Boros F, Maszlag-Török R, Vécsei L, Klivényi P (2020) Increased level of NEAT1 long non-coding RNA is detectable in peripheral blood cells of patients with Parkinson’s disease. Brain res. 1730:146672
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al (2010) Long noncoding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464(7291):1071
Liu S, Cui B, Dai Z, Shi P, Wang Z, Guo Y (2016) Long non-coding RNA HOTAIR promotes Parkinson’s disease induced by MPTP through up-regulating the expression of LRRK2. Curr Neurovasc Res 13(2):115
Zhou F, Xie S, Li J, Duan S (2019) Long noncoding RNA HOTAIR promotes cell apoptosis by sponging miR-221 in Parkinson’s disease. RSC Adv 9(51):29502–29510
Lang Y, Li Y, Yu H, Lin L, Chen X, Wang S et al (2020) HOTAIR drives autophagy in midbrain dopaminergic neurons in the substantia nigra compacta in a mouse model of Parkinson’s disease by elevating NPTX2 via miR-221-3p binding. Aging (Albany NY) 12(9):7660
Zhang Q, Huang X-M, Liao J-X, Dong Y-K, Zhu J-L, He C-C, et al. LncRNA HOTAIR promotes neuronal damage through facilitating NLRP3 mediated-pyroptosis activation in Parkinson’s disease via regulation of miR-326/ELAVL1 axis. Cell mol neurobiol.
Lin Q, Hou S, Dai Y, Jiang N, Lin Y (2019) LncRNA HOTAIR targets miR-126-5p to promote the progression of Parkinson’s disease through RAB3IP. Biol Chem 400(9):1217
Yang L, Zhang X, Li H, Liu J (2016) The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol BioSyst 12(8):2605
Zhang Q, Huang X-M, Liao J-X, Dong Y-K, Zhu J-L, He C-C, et al. (2020) LncRNA HOTAIR promotes neuronal damage through facilitating NLRP3 mediated-pyroptosis activation in Parkinson’s disease via regulation of miR-326/ELAVL1 axis. Cellular and Molecular Neurobiology:1–14
Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z et al (2010) A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29(18):3082
Bennett M (2005) The role of alpha-synuclein in neurodegenerative diseases. Pharmacol Ther 105(3):311
Dehay B, Bourdenx M, Gorry P, Przedborski S, Vila M, Hunot S et al (2015) Targeting α-synuclein for treating Parkinson’s disease: mechanistic and therapeutic considerations. The Lancet Neurology 14(8):855
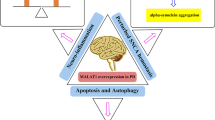
Cai L-J, Tu L, Huang X-M, Huang J, Qiu N, Xie G-H et al (2020) LncRNA MALAT1 facilitates inflammasome activation via epigenetic suppression of Nrf2 in Parkinson’s disease. Mol Brain 13(1):130
Sarkar S, Malovic E, Harishchandra DS, Ghaisas S, Panicker N, Charli A, et al. (2017) Mitochondrial impairment in microglia amplifies NLRP3 inflammasome proinflammatory signaling in cell culture and animal models of Parkinson’s disease. NPJ Parkinson’s Disease 3
Gureev A, Popov V (2019) Nrf2/ARE pathway as a therapeutic target for the treatment of Parkinson diseases. Neurochem Res 44(10):2273
Okorji U, Velagapudi R, El-Bakoush A, Fiebich B, Olajide O (2016) Antimalarial drug artemether inhibits neuroinflammation in BV2 microglia through Nrf2-dependent mechanisms. Mol Neurobiol 53(9):6426
Chen Q, Huang X, Li R (2018) lncRNA MALAT1/miR-205-5p axis regulates MPP+-induced cell apoptosis in MN9D cells by directly targeting LRRK2. Am J Trans Res 10(2):563
Esteves AR, Swerdlow RH, Cardoso SM (2014) LRRK2, a puzzling protein: insights into Parkinson’s disease pathogenesis. Experimental neurology 206
Imai Y, Gehrke S, Wang H-Q, Takahashi R, Hasegawa K, Oota E et al (2008) Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J 27(18):2432
Lee B, Shin J, VanKampen J, Petrucelli L, West A, Ko H et al (2010) Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson’s disease. Nat Med 16(9):998
Liu W, Zhang Q, Zhang J, Pan W, Zhao J, Xu Y (2017) Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell & Bioscience 7
Majidinia M, Mihanfar A, Rahbarghazi R, Nourazarian A, Bagca B, Avci ÇB (2016) The roles of non-coding RNAs in Parkinson’s disease. Mol Biol Rep 43(11):1193–1204
Author information
Authors and Affiliations
Contributions
Idea development: Eskandar Taghizadeh; literature search: Eskandar Taghizadeh and Forough Taheri; drafting: Seyed Mohammad Gheibihayat and Mohammadreza Afshani; critical revisions: Alihossein Saberi.
Corresponding authors
Ethics declarations
Ethical approval
None.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Taghizadeh, E., Gheibihayat, S.M., Taheri, F. et al. LncRNAs as putative biomarkers and therapeutic targets for Parkinson’s disease. Neurol Sci 42, 4007–4015 (2021). https://doi.org/10.1007/s10072-021-05408-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-021-05408-7