Abstract
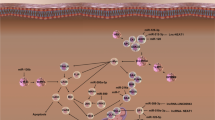
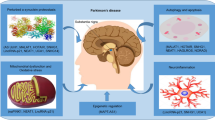
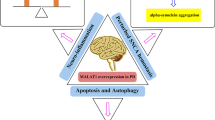
Parkinson’s disease (PD) is considered as a high prevalence neurodegenerative disorders worldwide. Pathologically, the demise of dopamine-producing cells, in large part due to an abnormal accumulation of the α-synuclein in the substantia nigra, is one of the main causes of the disease. Up until now, many de novo investigations have been conducted to disclose the mechanisms underlying in PD. Among them, impacts of non-coding RNAs (ncRNAs) on the pathogenesis and/or progression of PD need to be highlighted. microRNAs (miRNAs) and long ncRNAs (lncRNAs) are more noteworthy in this context. miRNAs are small ncRNAs (with 18–25 nucleotide in length) that control the expression of multiple genes at post-transcriptional level, while lncRNAs have longer size (over 200 nucleotides) and are involved in some key biological processes through various mechanisms. Involvement of miRNAs has been well documented in the development of PD, particularly gene expression. Hence, in this current review, we will discuss the impacts of miRNAs in regulation of the expression of PD-related genes and the role of lncRNAs in the pathogenesis of PD.

Similar content being viewed by others
References
Wu Y, Le W, Jankovic J (2011) Preclinical biomarkers of Parkinson disease. Arch Neurol 68(1):22–30
Zheng B et al (2010) PGC-1alpha, a potential therapeutic target for early intervention in Parkinson’s disease. Sci Transl Med . doi:10.1126/scitranslmed.3001059
Papapetropoulos S, McCorquodale D (2007) Gene-expression profiling in Parkinson’s disease: discovery of valid biomarkers, molecular targets and biochemical pathways. Future Neurol 2:29–38
Hamza TH et al (2010) Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat Genet 42(9):781–785
Ibanez P et al (2004) Causal relation between α-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 364(9440):1169–1171
Chiba-Falek O, Lopez GJ, Nussbaum RL (2006) Levels of α-synuclein mRNA in sporadic Parkinson disease patients. Mov Disord 21(10):1703–1708
Mandel S et al (2007) Applying transcriptomic and proteomic knowledge to Parkinson’s disease drug discovery. Exp Opin Drug Disc 2(9):1225–1240
Grünblatt E (2012) Parkinson’s disease: molecular risk factors. Parkinsonism Relat Disord 18:S45–S48
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12(12):861–874
Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6(4):259–269
Hammond SM (2005) MicroRNAs as tumor suppressors. Nat Genet 39(5):582–583
Croce CM (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10(10):704–714
Nicoloso MS et al (2009) MicroRNAs—the micro steering wheel of tumour metastases. Nat Rev Cancer 9(4):293–302
Salta E, De Strooper B (2012) Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol 11(2):189–200
Ambros V (2001) microRNAs: tiny regulators with great potential. Cell 107(7):823–826
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854
Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318(5858):1931–1934
Hu HY et al (2012) Evolution of the human-specific microRNA miR-941. Nature Commun 3:1145
Robles AI, Harris CC (2013) A primate-specific microRNA enters the lung cancer landscape. Proc Natl Acad Sci 110(47):18748–18749
Pauli A, Rinn JL, Schier AF (2011) Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12(2):136–149
Wang X et al (2011) The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol 3:a003756
Gupta RA et al (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464(7291):1071–1076
Guttman M et al (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458(7235):223–227
Pauli A et al (2012) Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22(3):577–591
Li X et al (2013) Long noncoding RNAs: insights from biological features and functions to diseases. Med Res Rev 33(3):517–553
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10(3):155–159
Mercer TR et al (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci 105(2):716–721
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136(4):629–641
Managadze D et al (2011) Negative correlation between expression level and evolutionary rate of long intergenic noncoding RNAs. Genome Biol Evol 3:1390–1404
Melton C, Judson RL, Blelloch R (2010) Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463(7281):621–626
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233
Ameres SL, Zamore PD (2013) Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 14(8):475–488
Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11(9):597–610
Rüegger S, Großhans H (2012) MicroRNA turnover: when, how, and why. Trends Biochem Sci 37(10):436–446
Westholm JO, Lai EC (2011) Mirtrons: microRNA biogenesis via splicing. Biochimie 93(11):1897–1904
Fabian MR, Sonenberg N (2012) The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 19(6):586–593
Gregory RI et al (2005) Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123(4):631–640
Schwarz DS et al (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115(2):199–208
Griffiths-Jones S (2004) The microRNA registry. Nucleic Acids Res 32(suppl 1):D109–D111
Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9(2):102–114
Chen G et al (2013) LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 41(D1):D983–D986
Khalil AM et al (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106(28):11667–11672
Guttman M et al (2010) Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28(5):503–510
Dinger ME et al (2008) Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18(9):1433–1445
Katayama S et al (2005) Antisense transcription in the mammalian transcriptome. Science 309(5740):1564–1566
Faghihi MA, Wahlestedt C (2009) Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10(9):637–643
He Y et al (2008) The antisense transcriptomes of human cells. Science 322(5909):1855–1857
Werner A, Sayer JA (2009) Naturally occurring antisense RNA: function and mechanisms of action. Curr Opin Nephrol Hypertens 18(4):343–349
Rastinejad F, Blau HM (1993) Genetic complementation reveals a novel regulatory role for 3′ untranslated regions in growth and differentiation. Cell 72(6):903–917
Sanchez-Herrero E, Akam M (1989) Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development 107(2):321–329
Heintzman ND et al (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39(3):311–318
Visel A et al (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457(7231):854–858
Kim T-K et al (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465(7295):182–187
Tisseur M, Kwapisz M, Morillon A (2011) Pervasive transcription–lessons from yeast. Biochimie 93(11):1889–1896
Mattick JS (2010) Linc-ing long noncoding RNAs and enhancer function. Dev Cell 19(4):485–486
Jalali S et al (2013) Systematic transcriptome wide analysis of lncRNA–miRNA interactions. PLoS One 8(2):e53823
Ouellet DL et al. (2006) MicroRNAs in gene regulation: when the smallest governs it all. BioMed Research International
Friedman RC et al (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1):92–105
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. cell 120(1):15–20
Hutvágner G, Zamore PD (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science 297(5589):2056–2060
Zeng Y, Cullen BR (2003) Sequence requirements for micro RNA processing and function in human cells. RNA 9(1):112–123
Zeng Y, Wagner EJ, Cullen BR (2002) Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell 9(6):1327–1333
Doench JG, Petersen CP, Sharp PA (2003) siRNAs can function as miRNAs. Genes Dev 17(4):438–442
Seggerson K, Tang L, Moss EG (2002) Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Developmental biology 243(2):215–225
Brennecke J et al (2003) Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113(1):25–36
Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21(6):354–361
Bernstein E, Allis CD (2005) RNA meets chromatin. Genes Dev 19(14):1635–1655
Whitehead J, Pandey GK, Kanduri C (2009) Regulation of the mammalian epigenome by long noncoding RNAs. Biochimica et Biophysica Acta 1790(9):936–947
Bracken AP, Helin K (2009) Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 9(11):773–784
Guenther MG et al (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130(1):77–88
Bonasio R, Tu S, Reinberg D (2010) Molecular signals of epigenetic states. Science 330(6004):612–616
Hung T, Chang HY (2010) Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol 7(5):582–585
Lee JT (2009) Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev 23(16):1831–1842
Spitale RC, Tsai M-C, Chang HY (2011) RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics 6(5):539–543
Good MC, Zalatan JG, Lim WA (2011) Scaffold proteins: hubs for controlling the flow of cellular information. Science 332(6030):680–686
Cheng L-C, Tavazoie M, Doetsch F (2005) Stem cells: from epigeneticsto microRNAs. Neuron 46(3):363–367
Jin P, Alisch RS, Warren ST (2004) RNA and microRNAs in fragile X mental retardation. Nat Cell Biol 6(11):1048–1053
Schratt GM et al (2006) A brain-specific microRNA regulates dendritic spine development. Nature 439(7074):283–289
Sempere LF et al (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5(3):R13
Smirnova L et al (2005) Regulation of miRNA expression during neural cell specification. Eur J Neurosci 21(6):1469–1477
Kosik KS, Krichevsky AM (2005) The elegance of the microRNAs: a neuronal perspective. Neuron 47(6):779–782
Krichevsky AM et al (2003) A microRNA array reveals extensive regulation of microRNAs during brain development. RNA 9(10):1274–1281
Chen W, Qin C (2015) General hallmarks of microRNAs in brain evolution and development. RNA Biol 12(7):701–708
Lopez JP et al (2014) miR-1202: a primate specific and brain enriched mirna involved in major depression and antidepressant treatment. Nat Med 20(7):764
John B et al (2004) Human microRNA targets. PLoS Biol 2(11):e363
Kim J et al (2004) Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci 101(1):360–365
Martin KC, Kosik KS (2002) Synaptic tagging—who’s it? Nat Rev Neurosci 3(10):813–820
Schaeffer C et al (2003) The RNA binding protein FMRP: new connections and missing links. Biol Cell 95(3–4):221–228
Lugli G et al (2005) Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem 94(4):896–905
Ashraf SI et al (2006) Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 124(1):191–205
Mehler MF, Mattick JS (2007) Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev 87(3):799–823
Qureshi IA, Mattick JS, Mehler MF (2010) Long non-coding RNAs in nervous system function and disease. Brain Res 1338:20–35
Dauer W, Przedborski S (2003) Parkinson’s disease: mechanisms and models. Neuron 39(6):889–909
Marsden C (1982) Neuromelanin and Parkinson’s disease. J Neural Transm Suppl 19:121–141
Wu D-C et al (2003) NADPH oxidase mediates oxidative stress in the 1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine model of Parkinson’s disease. Proc Natl Acad Sci 100(10):6145–6150
Uhl GR (1998) Hypothesis: the role of dopaminergic transporters in selective vulnerability of cells in Parkinson’s disease. Ann Neurol 43(5):555–560
Damier P et al (1993) Glutathione peroxidase, glial cells and Parkinson’s disease. Neuroscience 52(1):1–6
Dexter D et al (1989) Increased nigral iron content and alterations in other metal ions occurring in brain in Parkinson’s disease. J Neurochem 52(6):1830–1836
Riederer P et al (1989) Transition metals, ferritin, glutathione, and ascorbic acid in parkinsonian brains. J Neurochem 52(2):515–520
Sofic E et al (1992) Reduced and oxidized glutathione in the substantia nigra of patients with Parkinson’s disease. Neurosci Lett 142(2):128–130
Blum D et al (2001) Molecular pathways involved in the neurotoxicity of 6-OHDA, dopamine and MPTP: contribution to the apoptotic theory in Parkinson’s disease. Prog Neurobiol 65(2):135–172
Klein C, Westenberger A (2012) Genetics of Parkinson’s disease. Cold Spring Harb Perspect Medicin 2(1):a008888
Farrer M et al (2004) Comparison of kindreds with parkinsonism and α-synuclein genomic multiplications. Ann Neurol 55(2):174–179
Singleton A et al (2003) α-Synuclein locus triplication causes Parkinson’s disease. Science 302(5646):841
West AB et al (2005) Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci USA 102(46):16842–16847
Brice A (2005) Genetics of Parkinson’s disease: lRRK2 on the rise. Brain 128(12):2760–2762
Heman-Ackah SM et al (2013) RISC in PD: the impact of microRNAs in Parkinson’s disease cellular and molecular pathogenesis. Frontiers in molecular neuroscience. doi:10.3389/fnmol.2013.00040
Lev N et al (2006) Role of DJ-1 in Parkinson’s disease. J Mol Neurosci 29(3):215–225
Mizuno Y et al (2001) Parkin and Parkinson’s disease. Curr Opin Neurol 14(4):477–482
Dawson TM, Dawson VL (2010) The role of parkin in familial and sporadic Parkinson’s disease. Mov Disord 25(S1):S32–S39
Jones R (2010) The roles of PINK1 and Parkin in Parkinson’s disease. PLoS Biol 8(1):e1000299
Gandhi S et al (2006) PINK1 protein in normal human brain and Parkinson’s disease. Brain 129(7):1720–1731
Dehay B et al (2012) Lysosomal dysfunction in Parkinson disease: aTP13A2 gets into the groove. Autophagy 8(9):1389–1391
Park JS. et al (2014) Parkinson’s disease-associated human ATP13A2 (PARK9) deficiency causes zinc dyshomeostasis and mitochondrial dysfunction. Human mole genet p ddt623
Kim J et al (2007) A MicroRNA feedback circuit in midbrain dopamine neurons. Science 317(5842):1220–1224
Doxakis E (2010) Post-transcriptional regulation of α-synuclein expression by mir-7 and mir-153. J Biol Chem 285(17):12726–12734
Junn E et al (2009) Repression of α-synuclein expression and toxicity by microRNA-7. Proc Natl Acad Sci 106(31):13052–13057
Wang G et al (2008) Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of α-synuclein. Am J Hum Genet 82(2):283–289
Margis R, Margis R, Rieder CR (2011) Identification of blood microRNAs associated to Parkinsońs disease. J Biotechnol 152(3):96–101
Valadi H et al (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9(6):654–659
Cardo LF et al (2013) Profile of microRNAs in the plasma of Parkinson’s disease patients and healthy controls. J Neurol 260(5):1420
Soreq L et al (2013) Small RNA sequencing-microarray analyses in Parkinson leukocytes reveal deep brain stimulation-induced splicing changes that classify brain region transcriptomes. Front Mol Neurosci 6:10
Khoo SK et al (2012) Plasma-based circulating MicroRNA biomarkers for Parkinson’s disease. J Parkinson’s Dis 2(4):321–331
Gehrke S et al (2010) Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 466(7306):637–641
Cho HJ et al (2013) MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum Mol Genet 22(3):608–620
Shaked I et al (2009) MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity 31(6):965–973
Yang D et al (2012) miR-132 regulates the differentiation of dopamine neurons by directly targeting Nurr1 expression. J Cell Sci 125(7):1673–1682
Soreq H (2015) MicroRNA-target interactions in neurodegenerative diseases. SpringerPlus 4(Suppl 1):L1
Kanagaraj N et al (2014) Downregulation of miR-124 in MPTP-treated mouse model of Parkinson’s disease and MPP iodide-treated MN9D cells modulates the expression of the calpain/cdk5 pathway proteins. Neuroscience 272:167–179
Gong X et al (2016) miR-124 regulates cell apoptosis and autophagy in dopaminergic neurons and protects them by regulating AMPK/mTOR pathway in Parkinson’s disease. Am J Transl Res 8(5):2127–2137
Kim J et al (2015) MicroRNA-124 regulates glucocorticoid sensitivity by targeting phosphodiesterase 4B in diffuse large B cell lymphoma. Gene 558(1):173–180
Ledderose C et al (2012) Corticosteroid resistance in sepsis is influenced by microRNA-124–induced downregulation of glucocorticoid receptor-α*. Crit Care Med 40(10):2745–2753
Herrero, M.-T., et al., Inflammation in Parkinson’s disease: role of glucocorticoids. Front Neuroanat 2015. 9
Miñones-Moyano E et al (2011) MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet 20(15):3067–3078
Soreq L et al (2014) Long non-coding RNA and alternative splicing modulations in Parkinson’s leukocytes identified by RNA sequencing. PLoS Comput Biol 10(3):e1003517
Carrieri C et al (2015) Expression analysis of the long non-coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson’s disease. Front Cellular Neurosci 9:114
Matsui M, Corey DR (2016) Non-coding RNAs as drug targets. Nat Rev Drug Discov. doi:10.1038/nrd.2016.117
Bennett CF, Swayze EE (2010) RNA targeting therapeutics: molecular mechanisms ofAntisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol 50:259–293
Chen X (2015) Predicting lncRNA-disease associations and constructing lncRNA functional similarity network based on the information of miRNA. Sci Rep 17(5):13186
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors have declared no conflicts of interest.
Rights and permissions
About this article
Cite this article
Majidinia, M., Mihanfar, A., Rahbarghazi, R. et al. The roles of non-coding RNAs in Parkinson’s disease. Mol Biol Rep 43, 1193–1204 (2016). https://doi.org/10.1007/s11033-016-4054-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-016-4054-3