Abstract
The ecological success of tropical corals is regulated by symbiotic dinoflagellate algae (Symbiodiniaceae). Corals can associate with multiple Symbiodiniaceae species simultaneously, yet the conditions that permit Symbiodiniaceae cohabitation are not understood. We examined how corals self-shade their own tissues causing within-colony light gradients that drive Symbiodiniaceae photoacclimatory processes and positional genetic disparity. Paired light ‘exposed’ and ‘shaded’ samples from 20 coral species were collected from a shallow coral reef (Rarotonga, Cook Islands). Through active chlorophyll fluorometry, rapid light curves revealed that exposed Symbiodiniaceae exhibited 50% higher values in minimum saturating irradiances and demonstrated a shift towards preferential nonphotochemical quenching [1 – Q], consistent with higher overall light exposure. High-throughput or targeted DNA sequencing of ITS2 and psbAncr markers demonstrated that corals harboured distinct and/or differentially abundant Symbiodiniaceae ITS2 sequences (typically rare in relative abundance) or multiple ITS2 intragenomic variant profiles across shaded vs exposed regions. In Hydnophora cf. microconos, within-colony symbiont genetic disparity was positively correlated with the magnitude of difference in [1 – Q] utilisation. Together, these results suggest that within-colony light gradients produce distinct optical niches that enable symbiont cohabitation via photoadaptation, a phenomenon that is expected to increase the adaptive capacity of corals under future climates.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ecological success of tropical corals is strongly regulated by their mutualistic symbiosis with the Symbiodiniaceae, a family of photosynthetic dinoflagellate algae that live within coral endodermal cells (Muscatine 1990). Tight nutrient recycling between these partners supports the metabolic requirements for coral survival and growth (Muscatine 1990; Gattuso et al. 1999; Davy et al. 2012). The Symbiodiniaceae is a diverse family of 16 primary lineages, with 11 formally described as genera (LaJeunesse et al. 2018; Nitschke et al. 2020; Pochon and LaJeunesse 2021; Yorifuji et al. 2021). Symbiodiniaceae species have evolved different physiological optima for environmental variables (Rowan 2004; Abrego et al. 2008; Howells et al. 2012; Hoadley et al. 2021) and may confer traits (e.g. thermal tolerance) to their host (Berkelmans and Van Oppen 2006; Sampayo et al. 2008; Oliver and Palumbi 2011).
A coral may host multiple Symbiodiniaceae species simultaneously, typically with a single highly abundant ‘dominant symbiont’ and one or more low abundance ‘background symbionts’ (Mieog et al. 2007; Silverstein et al. 2012; Boulotte et al. 2016). Low abundance Symbiodiniaceae may also confer environmental stress resistance to their host by supplying a source of genomic innovation, enhancing the resilience of host–symbiont communities (Bay et al. 2016; Ziegler et al. 2018). Studies have proposed that corals may acclimatise or adapt to warmer oceans through rearrangements of symbiont community compositions favouring thermally tolerant species (Buddemeier and Fautin 1993; Baker et al. 2004; Berkelmans and Van Oppen 2006; Cunning et al. 2018; Claar et al. 2020), a phenomenon observed following temperature-induced coral bleaching (Berkelmans and Van Oppen 2006; LaJeunesse et al. 2009; Cunning et al. 2015; Oladi et al. 2021). However, some tolerant symbiont species are considered less compatible with the nutritional needs of their host (Stat and Gates 2011; Pettay et al. 2015; Matthews et al. 2017; Gabay et al. 2018; Sproles et al. 2020), and perturbed symbiont communities tend to return to their original composition over time if stress is alleviated (Thornhill et al. 2006; Stat et al. 2009b; McGinley et al. 2012). Temporary community shifts suggest a hierarchy to symbiont cohabitation (McIlroy et al. 2019, 2020), however the ecological processes (i.e. competition) governing Symbiodiniaceae species interactions have not been fully resolved, and the heterogeneously distributed resource(s) responsible for within-colony, multiple symbiont species cohabitation has yet to be determined.
Light gradients on coral reefs provide significant opportunity for specialisation of Symbiodiniaceae photobiological traits and species partitioning across particular light environments (Sampayo et al. 2007). Examples of depth-driven light gradients and zonation of Symbiodiniaceae species in depth-generalist coral species are common, leading to hypotheses that Symbiodiniaceae are partitioned into optical niches (Rowan and Knowlton 1995; Sampayo et al. 2007; Frade et al. 2008; Goulet et al. 2019). However, it is unknown if the same notion can be applied broadly to the light microenvironments that exist across individual coral colonies. Corals build morphologically heterogeneous structures that inevitably shelf-shade, producing pronounced light gradients within a single colony (Anthony et al. 2005; Kaniewska et al. 2008; Ow and Todd 2010) which, in turn, may create multiple niches for distinct symbiont species to exploit (Suggett et al. 2017). Spatial diversification of symbionts across individual colonies is evident for several coral species (Rowan and Knowlton 1995; van Oppen et al. 2001; Garren et al. 2006) and certain Symbiodiniaceae genera appear to display consistency regarding their preference for light ‘exposed’ or ‘shaded’ coral surfaces (Rowan and Knowlton 1995; Garren et al. 2006; Kemp et al. 2008). Localised or patchy bleaching of individual corals is common (Celliers and Schleyer 2002; Penin et al. 2007), and symbiont-retentive regions may correlate with the zonation of distinct Symbiodiniaceae species with superior tolerances of high temperatures and irradiance (Rowan et al. 1997; Kemp et al. 2014). Symbionts must make continuous phenotypic adjustments in response to variations in light intensity, referred to as photoacclimation. The upper and lower limits of photosynthetic optimisation (photoacclimatory potential) can vary considerably between different Symbiodiniaceae species (Iglesias-Prieto and Trench 1994, 1997; Reynolds et al. 2008) or even between sister strains of the same species (Suggett et al. 2007, 2015; Díaz-Almeyda et al. 2017). Studies of symbionts in the coral Goniastrea aspera demonstrated that shaded surfaces experience chronic photoinhibition have lower recovery potential and are more prone to bleaching when exposed to elevated irradiance and temperatures (Brown et al. 2000a, 2000b, 2002). As such, it is plausible that disparities in photoacclimatory potential between symbionts may be the trait responsible for structuring symbiont communities across coral colonies.
Here, we characterise Symbiodiniaceae photochemical signatures of high-light and low-light niches of individual coral colonies, and test whether the disparity between high-light and low-light acclimation correlates with Symbiodiniaceae genetic disparity and multiple symbiont cohabitation. We hypothesised that the dynamic range of optical niches will vary across coral species, and that disparities in photoacclimation states will correlate with symbiont community compositional changes across the colony surface.
Methods
Sampling location and coral collection
Over an eight-day period, 100 coral colonies were sampled from the shallow (< 2 m depth) lagoon of Rarotonga (September 2019, Cook Islands; ~ 21°12′7.92″S, 159°45′30.65"W). A diverse range of 20 coral species spanning 11 genera were selected according to their prevalence (Table 1; Supplementary Fig. 1, Supplementary table 1); these species spanned multiple colony morphologies, including branching (corymbose, digitate, hispidose, and closed), tabulate, massive, and encrusting growth forms (Veron 2000). Two small coral fragments (approximately 1.5 cm2) were collected using a chisel from each colony, one each from exposed and shaded regions (n = 200 total samples). Exposed and shaded regions were defined by their apparent light-exposed (i.e. upward facing at the colony apex) or shaded (i.e. underhanging or downwards facing) position. Following collection, samples were immediately transported to an on-site aquarium (30 L) where they were maintained overnight with regular exchange of lagoon seawater and constant aeration.
Light utilisation capacity of exposed versus shaded regions
The photoacclimation state of each “exposed” or “shaded” fragment was estimated from fluorescence-light response curves of PSII photochemistry (MacIntyre et al. 2002). Active chlorophyll fluorescence was used to generate a rapid light curve (RLC) for each sample (Warner et al. 2010; Nitschke et al. 2018). The RLCs were produced using a Diving Pulse Amplitude Modulated (PAM) fluorometer (Walz GmbH, Effeltrich, Germany. Settings: MI: 6, Gain: 6, SI: 12, SW: 0.8 s, LC-INT: 3). To control for natural diel variation in the photo-physiological parameters, coral samples were subjected to RLCs in the first hour of daylight, when PSII fluorescence yields are expected to be maximal (Hill and Ralph 2005). The use of RLCs was preferred in this study to ensure all measurements were taken within a narrow time-frame, and we interpret the findings within the limitations of the method and not as absolute rates of electron transport in photosynthesis (Warner et al. 2010; Nitschke et al. 2018). Each RLC consisted of variable chlorophyll fluorescence yield measurements taken at nine discrete time-steps. A low-light acclimated (c.a. 5 µmol photons m−2 s−1) measurement of the minimum and maximum fluorescence yields (Fo and Fm, respectively) initiated the RLC, followed by measurements under actinic light (F′ and Fm′) at eight 30 s intervals of increasing light intensity (61, 91, 126, 214, 320, 503, 801, and 1409 µmol photons m−2 s−1). These parameters were used to estimate the maximum (Fv/Fm = [Fm—Fo]/Fm) and effective (Fq′/Fm′ = [Fm′ − F′]/Fm′) photochemical efficiencies of the Symbiodiniaceae cells in each sample.
Symbiodiniaceae photoacclimate to specific light environments by changing their saturating irradiance (Ek), which represents the transition between the light-limited and the light-saturated states of PSII, accompanied by changes to the maximum photochemical efficiency of PSII (Fq'/Fm'(max)) which typically increases under low-light conditions (Anthony and Hoegh-Guldberg 2003; Hennige et al. 2009). To estimate these parameters, the PE curves were fitted to a model (Eq. 1) that describes Ek and Fq'/Fm' (max) (Hennige et al. 2008).
Photoacclimatory adjustments also include shifts in the reliance on photochemical quenching and nonphotochemical quenching pathways (Robison and Warner 2006; Hennige et al. 2009). To estimate capacity to dissipate excitation energy via fluorescence and pathways of energy transfer other than fluorescence, the extent of light-dependant photochemical quenching ([1 – C], Eq. 2) and nonphotochemical quenching ([1 – Q], Eq. 3) were calculated at the final step in the RLC (Nitschke et al. 2018), to capture the maximum extent of dynamic quenching for each sample as:
Following PAM measurements, diffuse reflectance measurements of each sample were measured with a USB4000 spectroradiometer (Ocean Optics) at 1 nm resolution (400 to 750 nm) to broadly characterise the sample optical properties (D’Angelo et al. 2008; Winters et al. 2009; Cohen and Dubinsky 2015). A fibre-optic cable attached to a pulsed-xenon lamp (PX-2, Ocean Optics) was used to deliver full-spectrum light, and reflectance delivered to the USB2000 measuring unit via a fibre bifurcation. Reference spectra of the light source (acquired from a tissue-free coral skeleton) and background noise (in darkness) were stored and each measurement was acquired with the optic positioned at a 45° angle to the coral surface, at approximately 5 mm distance. Three acquisitions were averaged to provide the final spectrum. Raw data were processed (wavelength limits: 400 nm—750 nm; locally weighted scatterplot smoothing with a parametrised span of 0.1), and the total diffuse reflectance, i.e. the sum of the relative reflectance between 400 and 750 nm, was calculated with the R package “pavo” (Maia et al. 2013).
DNA extraction, PCR amplification, and sequencing
After photobiology data collection, each sample was rinsed with 0.22 µm filtered seawater, blot-dried with paper towel, and transferred to a salt-saturated DMSO preservation buffer (Seutin et al. 1991). DNA was extracted from each sample using a CTAB-chloroform DNA extraction protocol (Cunning et al. 2016). Symbiodiniaceae diversity was determined through sequencing of the ITS2 region. The “ITSDino” (5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGAATTGCAGAACTCCGTG-3’) (Pochon et al. 2001) and “ITS2-Rev2” (5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCCTCCGCTTACTTATATGCTT-3’) (Stat et al. 2009a) primers (including Illumina adaptor sequences) were used to amplify the ITS2 region in PCR. The optimised cycling conditions were denaturation for 3 min at 95 °C, followed by 25 cycles of 30 s at 95 °C, 30 s at 55 °C, 30 s at 72 °C, and a final extension for 5 min at 72 °C. PCR amplicons were then cleaned with AMPure XP beads following the manufacturer’s protocol. Cleaned PCR amplicons were normalised to 2 ng/µl for a second indexing PCR reaction with a Nextera XT Library Prep kit (Illumina) with a dual indexing strategy. To create the final library for sequencing, 5 µl of each indexed sample was pooled and the final library cleaned, again using the AMPure XP Bead protocol. A Bioanalyzer trace and Qubit were used to measure the nanomolarity of the final library, and it was sequenced at 5 pM on a 500 cycle MiSeq run with 15% PhiX spike-in.
To further explore Symbiodiniaceae diversity, the chloroplast psbA non-coding region (psbAncr) was amplified (a marker that carries phylogenetic resolution at the species to sub-species level in Symbiodiniaceae (LaJeunesse and Thornhill 2011; Thornhill et al. 2014)) for a subset of Acropora and Psammocora samples. Samples were selected based on the criterion of harbouring a single ITS2 profile (see below) in high abundance (> 80% in relative abundance of the sample total sequence count). The 7.4-For (5’-GCATGAAAGAAATGCACACAACTTCCC-3’) and 7.8-Rev (5’-GGTTCTCTTATTCCATCAATATCTACTG-3’) primer pair was used as previously described (LaJeunesse and Thornhill 2011), and PCR products were Sanger sequenced in both directions (Macrogen, South Korea).
Bioinformatics
Primer sequences were removed from all reads using Cutadapt (Martin 2011) and the raw sequence data were denoised using the DADA2 pipeline to delineate sequencing errors from biological diversity (Callahan et al. 2016) following the author-recommended ITS2 workflow. DADA2 ‘amplicon sequence variants’ (ASVs) were assigned a taxonomic identity using the RDP classifier (Wang et al. 2007) against a custom, previously published Symbiodiniaceae-enriched ITS2 reference database (github.com/nitschkematthew/Symbiodatabaceae). The default RDP bootstrap value of 50 was used to assign taxonomic ranks to an ASV; below this threshold the rank was set to ‘unassigned’. Non-Symbiodiniaceae sequences were removed at this point prior to any further analyses. To visualise the genetic diversity within Cladocopium across all samples, an alignment-free phylogenetic tree was generated using a k-mer (k = 7)-based clustering method [kmer, kdistance (Wilkinson 2018)].
To investigate Symbiodiniaceae diversity at a resolution that incorporates ITS2 intragenomic variation, the raw sequences (which are available online at the National Center for Biotechnology Information, Sequence Read Archive: BioProject number PRJNA863925) were submitted to SymPortal (Hume et al. 2019). SymPortal uses a minimum entropy decomposition (MED)-based approach to consolidate ITS2 sequences into nodes and catalogues novel post-MED sequences within the SymPortal ITS2 reference database. By comparing the assemblage of post-MED sequences between each sample, SymPortal identifies co-occurring ‘defining’ intragenomic sequence variants (DIVs) which are used to designate ITS2 type profiles representative of putative Symbiodiniaceae taxa (Hume et al. 2019).
To validate the identification of the closely-related ITS2 profiles within the C3 and C1 radiations of Cladocopium, the chloroplast psbAncr marker was analysed on a subset of samples. The psbAncr chromatograms were visually inspected and trimmed according to the stretch of unambiguous base calls. Samples with any low-quality sequences, potentially due to intra- or inter-genomic variation, were discarded. Due to the length of psbAncr amplicons, the forward and reverse reads did not overlap for the generation of consensus sequences. Therefore, the forward and reverse reads for each sample were aligned separately [DECIPHER, Alignseqs (Wright 2016)] and then concatenated before a maximum parsimony tree was produced using RAxML (Stamatakis 2014) on the CiPRES portal (HPC v.8 on XSEDE (Miller et al. 2011)) using default settings. FASTA files containing the concatenated forward and reverse complement psbA sequences can be retrieved from the supplementary material.
Statistical analyses
To compare the photo-physiological characteristics and reflectance spectra of symbionts found within shaded and exposed microhabitats, a Kruskal–Wallis test (nonparametric ANOVA) was performed on Fq′/Fm′(max), Ek, [1 – C], [1 – Q], and total diffuse reflectance between microhabitats from samples pooled across all coral species.
To explore ITS2 sequence differentiation between shaded and exposed microhabitats, ASV accumulation curves were produced for exposed samples only, and then on paired (within-colony) shaded and exposed samples with pooled ASVs [Vegan, specaccum (Dixon 2003)]. Across all coral samples, the number of ASVs that were detected in only shaded samples, only exposed samples, and in both microhabitats were counted. DeSeq2 was used to test for the differential abundance of ASVs and post-MED sequences between shaded and exposed habitats (Love et al. 2014), with a Wald test on the dispersion around group means. To control the false discovery rate, the p-values for all differential abundance tests were adjusted together using the Benjamini–Hochberg method. To prevent multiple observations of nil sequence abundance from significantly skewing the group means, the DeSeq2 results were filtered to retain only ASVs and post-MED sequences that were detected in at least four samples within at least one habitat. A permutational multivariate analysis of variance (PERMANOVA) was performed on ITS2 profiles across the dataset to test for differences between coral species and habitats [Vegan, adonis (Dixon 2003)].
Comparisons of symbiont community diversity using the ITS2 marker are complicated by the fact that unique ITS2 ASVs are not necessarily equivalent to distinct species. Therefore, to capture between-sample sequence diversity, we utilised the weighted UniFrac method to provide a phylogenetically-informed distance metric (Lozupone and Knight 2005). Coral species that had at least one differentially abundant sequence (i.e. significant within-colony genetic heterogeneity) were selected for further analyses. A phylogenetic tree was generated for ASV sequences using k-mer clustering [kmer, kdistance (Wilkinson 2018)] and a weighted UniFrac distance matrix was computed in phyloseq for each coral species [phyloseq, distance, McMurdie and Holmes (2013)]. For each coral species, a linear regression between the within-colony weighted UniFrac distance values and the within-colony fold difference in functional photobiological parameters (Ek, Fq′/Fm′(max), [1 – C], [1 – Q]) was performed. All linear regressions were corrected for multiple tests using the Benjamini–Hochberg method. From the DeSeq2 outputs and SymPortal ITS2 data, it was apparent that sequence abundance differences between optical habitats are common for rare sequences. Therefore, each sample was filtered for ASVs that represented < 1% of the total sample read count (a threshold used to define the “rare microbial biosphere”; (Boulotte et al. 2016)), and the weighted UniFrac distances between sample pairs were again correlated with the fold difference in photobiological parameters. All code required to reproduce data processing and analysis can be found at https://github.com/Robert-Lewis-NZ/Rarotonga_2019_shaded_vs_exposed.
Results
Photophysiology
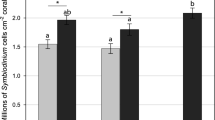
Photosynthetic acclimation states differed between shaded and exposed habitats for all parameters: Fq′/Fm′(max) (χ2 = 80.971, p < 0.001); Ek (χ2 = 43.07, p < 0.001); [1 – C] (χ2 = 68.74, p < 0.001); [1 – Q] (χ2 = 49.7, p < 0.001); and total diffuse reflectance (χ2 = 71.9, p < 0.001; Fig. 1). On average, shaded samples exhibited 13% higher Fq′/Fm′(max) and 50% lower Ek values than exposed samples; however, the fold differences varied considerably across the coral species (for species specific parameter estimates, see Supplementary Figs. 2–5). The mean maximal photochemical quenching coefficient [1 – C] of shaded samples was 47% lower than exposed samples, indicating greater reliance on photochemical quenching. Correspondingly, the mean maximal nonphotochemical coefficient [1 – Q] of shaded samples was 16% higher than exposed samples, indicating less reliance on nonphotochemical quenching. The mean total diffuse reflectance of exposed samples was 70% greater than shaded samples (Fig. 1).
Signatures of low and high light acclimation of Symbiodiniaceae across all coral species. Boxplots of A Maximum photochemical efficiency of PSII (Fq′/Fm′(max)) values and B minimum saturating irradiance (Ek) in shaded (grey) and exposed (white) samples. C) Mean maximal photochemical quenching [1 – C] and nonphotochemical quenching [1 – Q] coefficients of shaded (black symbol) and exposed (white symbol) samples (± 95% CI). The [1 – C]:[1 – Q] = 1 slope is depicted as a solid line. D) Mean reflectance of shaded (grey) and exposed (white) samples between 400 and 750 nm (dashed line above and below mean represent ± 95% CI). N = 98 per microhabitat type in all plots, integrating all coral species
Symbiodiniaceae ITS2 sequence diversity
Illumina ITS2 sequencing yielded 9,136,599 Symbiodiniaceae sequences after quality filtering and the removal of chimeric sequences. Two sample pairs (one from Pocillopora cf. damicornis and one from Echinopora sp.) were removed from the dataset due to low read counts. The total number of reads per sample averaged 46,615 ± 17,472 (SD). A total of 909 Symbiodiniaceae ASVs were assigned to four Symbiodiniaceae genera: Cladocopium, Durusdinium, Symbiodinium, and Breviolum, which were present within the dataset at a relative frequency of 0.856, 0.144, 0.0004, and 0.0001 of the total reads, respectively. The majority of ASVs were assigned to seven distinct radiations within Cladocopium (C1, C1c, C42, C15, C3, C3*, and C3k; Supplementary Fig. 6). Within-colony pooling of shaded and exposed sequences together provided 155 more distinct ASVs compared with exposed samples only across the community of 20 coral species (Fig. 2).
Symbiodiniaceae ITS2 sequence differences between shaded and exposed samples. A Accumulation curves of Symbiodiniaceae ITS2 (ASV) sequences for paired (within-colony) shaded and exposed samples pooled together (dashed line) and exposed samples only (solid line). The difference between the accumulation curves represents increasing evidence for symbiont niche differentiation as more colonies are sampled. B The total number of ITS2 ASVs exclusively detected in shaded samples only (left), exposed samples only (right), and in both microhabitats (centre)
Differential abundance of ITS2 sequences
No single ASV was found to be significantly differentially abundant in either habitat when all samples were pooled together, likely due to high levels of host-specificity. However, when analysed within each coral species, nine ASVs and seven post-MED sequences were significantly differentially abundant with two ASVs being consistently more abundant within exposed habitats across three coral species (Fig. 3). For most species, the differentially abundant ASVs were rare in relative abundance, except for Porites cf. lutea, where differentially abundant ASVs comprised up to 80.6% of the total reads. Excluding Porites cf. lutea, the mean relative frequency of all differentially abundant ASVs and post-MED sequences collectively pooled within each species was 0.031 ± 0.035 and 0.033 ± 0.056 (SD), respectively. Three (43%) of the differentially abundant post-MED sequences were DIVs within Cladocopium ITS2 profiles, and four (57%) were not DIVs in this dataset or any previous datasets within the SymPortal database (at the time of analysis).
Differential abundance of Symbiodiniaceae ITS2 sequences between shaded and exposed samples within each coral species. Log2 fold change in ASV abundance for significantly differentially abundant A DADA2 ASVs and B SymPortal post-MED sequences that are more abundant in exposed (square symbols) or shaded (circle symbols) samples (Benjamini–Hochberg adjusted p < 0.05)
ITS2 profile diversity: SymPortal analysis
SymPortal yielded a total of 8,991,072 post-MED Symbiodiniaceae sequences across all samples and assigned these sequences to the genera Cladocopium, Durusdinium, Symbiodinium and Breviolum. From these sequences, 31 ITS2 DIV profiles were generated, of which DIVs represented 83.4 ± 7.7% (SD) of the sequence counts within each sample (Fig. 4). PERMANOVA analyses of ITS2 profile composition revealed that Symbiodiniaceae assemblages varied significantly by species (F = 9.16, p = 0.001), but neither habitat nor the interaction between species and habitat had a significant effect on profile composition.
ITS2 DIV profiles of exposed and shaded coral samples. Pairs of horizontal bars represent matching exposed (left) and shaded (right) samples from a single coral colony. Each colour indicates a unique ITS2 DIV profile inferred by SymPortal. Symbiodiniaceae sequences not assigned to the sample profiles are depicted in grey
However, several coral-specific patterns in the distribution of ITS2 profiles were evident. All samples from each of Hydnophora cf. exesa, Galaxea cf. fascicularis, and Psammocora contigua, regardless of habitat, contained one profile (D1-D4-D4c-D6-D1d-D2, D1-D4-D4c-D1c-D2, and C1-C1b-C1c-C42.2-C1br-C1bh-C1cb-C72k, respectively). Four DIV profiles from the C3 radiation were found to be randomly distributed across the Acropora spp. in both habitats, all of which were also found in Hydnophora cf. microconos, Astrea curta, Echinopora sp., and Leptoria phrygia. When the C3f/C3fl-C3ae-C3-C3bj-C50a-C3h profile was observed in A. curta and Acropora cf. globiceps, it was dominant in both shaded and exposed habitats. Additional C3 majority sequence profiles not found in Acropora were present in Echinopora sp. and Leptastrea purpurea. Six profiles from the C42 radiation were found in the pocilloporids, with each colony dominated by a single profile.
Psammocora sp. 2, L. purpurea, Pavona cf. duerdeni, Echinopora sp., and Hydnophora cf. microconos each exhibited within-colony differentiation of ITS2 profiles. Two C1 majority sequence profiles dominated the colonies of P. contigua and alternated between exposed and shaded depending on the colony. One L. purpurea colony contained three distinct profiles; C3 and C1 majority sequence profiles co-dominated the exposed sample, while a distinct C1 majority sequence profile dominated the shaded sample. Two P. duerdeni colonies harboured two and three distinct profiles each; C1 majority sequence profiles dominated the exposed samples, whereas a single DIV profile (C21) occupied the shaded samples of both colonies. Half of the E. gemmacea colonies had differentiated profile compositions; C3 and D1 majority sequence profiles dominated the exposed and shaded samples of one colony, respectively, whilst this pattern was reversed in the other colony. Four H. microconos colonies harboured two or three distinct profiles per colony; C3 majority sequence profiles were more abundant in exposed samples, whereas C1 and/or D1 majority sequence profiles dominated the shaded samples (Fig. 4).
psbA ncr sequence diversity
The psbAncr alignments for samples containing C3 or C1 ITS2 majority sequence profiles contained multiple blocks of indels. Phylogenetic analysis of nucleotide substitutions revealed that the C3f/C3fl-C3ae-C3-C3bj-C50a-C3h profile formed a divergent clade to the other C3 majority sequence profiles found in Acropora (Supplementary Fig. 7), consistent with C3f/C3fl-C3ae-C3-C3bj-C50a-C3h co-occurrence in both exposed and shaded samples of single colonies. The two C1 majority sequence profiles (C1-C1b-C1c-C42.2-C1br-C1bh-C1cb-C72k and C1/C3-C1c-C1b-C72k-C1w) were not congruent with psbAncr diversity.
ITS 2 sequence diversity and photo-physiology
When analysed on the complete sample ASV diversity, no linear regressions between the paired-sample difference in the photobiological parameters and UniFrac distance were significant. However, after filtering for only rare sequences (< 1%) within each sample, H. cf. microconos displayed a strong positive correlation between the paired-sample difference in extent of [1 – Q] and the ASV UniFrac distances (Fig. 5), suggesting that rare sequences are diagnostic of functionally distinct symbiont communities or genotypes within a single colony.
Photosynthetic signatures that correlate with Symbiodiniaceae genetic disparity. Within-colony difference in [1 – Q] utilisation versus within-colony genetic disparity (weighted unifrac distance) of rare (< 1%) Symbiodiniaceae sequences in Hydnophora cf. microconos. Points represent individual H. cf. microconos colonies. The line represents the linear regression fit to data (F = 11.2, Benjamini–Hochberg adjusted p = 0.042)
Discussion
Five distinct but interrelated findings are presented here which provide insights into within-colony partitioning of Symbiodiniaceae. First, symbiont communities inhabiting shaded and light exposed coral tissues consistently demonstrate photobiological signatures of acclimation or adaptation to ‘low’ and ‘high’-light exposure, respectively. Second, pooling shaded and exposed samples yielded an additional 155 ITS2 sequences compared to exposed samples alone, suggesting this sampling design can serve as a new standard for the retrieval of Symbiodiniaceae genetic diversity. Third, multiple coral species have differentially abundant background Symbiodiniaceae ITS2 sequences, suggesting that the rare symbiont biosphere may exhibit substantial community differentiation between optical niches, or that rare sequence variants may be diagnostic of strain-level partitioning. Fourth, multiple coral species exhibited distinct ITS2 profiles in shaded versus exposed regions of individual colonies, suggesting that symbionts competing within a colony can become locally dominant via optical niche partitioning, and this may be more common on coral reefs than realised. Fifth, for Hydnophora microconos, the within-colony UniFrac distances of rare (< 1%) Symbiodiniaceae sequences were strongly positively correlated with the difference in [1 – Q] utilisation, suggesting that differences in functional capacity may underpin symbiont community differentiation. These lines of evidence support the notion that within-colony light gradients influence Symbiodiniaceae species cohabitation, highlighting a need for an approach to symbiont ecology from the perspective of coral colonies as heterogenous habitats.
Shaded and exposed tissues yield distinct and differentially abundant sequences
Dividing coral colonies into regions defined as shaded or light exposed revealed Symbiodiniaceae genetic disparity. In most cases, the collective abundance of the differentially abundant sequences was relatively low, suggesting that rare symbionts are regulated by light availability to a greater extent than dominant symbionts. Although background Symbiodiniaceae sequences are frequently detected in deep-sequencing analyses (Arif et al. 2014; Quigley et al. 2014; Thomas et al. 2014; Hume et al. 2020), the factors regulating the prevalence and diversity of rare symbionts remain largely unresolved (Boulotte et al. 2016; Lee et al. 2016). The limited research to date suggests that rare symbionts may be more sensitive or responsive to environmental change than dominant symbiont species. For example, Boulotte et al. (2016) profiled the ITS2 diversity in two coral species before and after two separate bleaching events and found that novel rare Symbiodiniaceae sequences were detected after each bleaching episode. Similarly, Lee et al. (2016) found that the presence of background Symbiodiniaceae appeared to fluctuate substantially throughout an annual cycle. Differentially abundant sequences could also represent strain-level intragenomic variation among dominant symbionts (LaJeunesse et al. 2010; Arif et al. 2014), with each strain occupying a different region in the colony. Most (57%) of the differentially abundant post-MED sequences of this study did not belong to an ITS2 profile and were not DIVs of any other ITS2 profiles in the SymPortal reference database, suggesting that these environmentally responsive Symbiodiniaceae species (or intragenomic variants) are consistently rare. Assigning these rare sequences to genomes and developing technologies to visualise these cells within coral tissues (e.g. via fluorescence in situ hybridisation (Yokouchi et al. 2003)) will be essential to measure the functional significance of their physiologies and the adaptations that enable them to persist in the presence of abundant competitors.
Symbiont cohabitation across microhabitats via the analysis of ITS2 profiles
Several coral species, including H. cf. microconos, P. cf. duerdeni, Echinopora sp., and L. purpurea, displayed marked within-colony differences in the dominant ITS2 profiles, including examples of both Cladocopium-Cladocopium and Cladocopium-Durusdinium cohabitation. Within-colony partitioning of Symbiodiniaceae species into optical niches where they become the locally dominant symbiont may be more common in coral communities than previously realised. However, within these examples, the dominant symbiont community was not always differentiated by light exposure. Competitive hierarchies and successional processes may cause adult colonies to become almost entirely dominated by a single competitively-superior symbiont species (Abrego et al. 2009; McIlroy et al. 2019), despite the potential for cohabitation with additional symbiont species. Although competitive hierarchies may depend on the prevailing light conditions (McIlroy et al. 2019), it seems plausible that coral-symbiont coevolution may produce partnerships that function optimally under a wide range of conditions (Gabay et al. 2019). In turn, these optimal partners may exclude other symbiont species from establishing, producing a relatively homogeneous symbiont community among individuals of a given coral species.
Morphological variation between colonies of the same species may cause within-colony light gradients to be more pronounced for some individuals (as evidenced by the variation in the fold differences of photobiological parameters within each species; Supplementary Figs. 2 and 3). It is notable that the four coral species with differentiated symbiont communities had either massive or encrusting morphologies, and spatial differentiation of symbionts has been observed among these growth forms in past studies (Garren et al. 2006; Kemp et al. 2008, 2014). These skeletal morphologies may be more likely to form overhanging protrusions or cervices and self-shade compared to branching morphologies which are considered more effective at redistributing light throughout the colony via diffuse scattering compared to massive skeletons (Marcelino et al. 2013; Enríquez et al. 2017). Ultimately, establishing a causal link between coral bio-optics and symbiont niche differentiation will require a quantitative spatial framework that integrates the three-dimensional complexity of individual corals and the corresponding gradients of incident light.
psbA ncr sequences hint at Cladocopium strain-level differentiation
Several psbAncr Cladocopium haplotypes were recovered (containing substitutions and/or insertions/deletions) that were not congruent with the corresponding ITS2 DIV profile. This may be indicative of strain-level genetic variation in which an assortment of Cladocopium strains of a single species are distributed heterogeneously between and within coral colonies (see also Thornhill et al. 2014). However, psbAncr haplotypes assigned to the C3f/C3fl-C3ae-C3-C3bj-C50a-C3h and C3f-C3fl-C50a-C3-C3ae-C3fm-C3h-C3bj ITS2 DIV profiles formed distinct clades, potentially representing a distinct, but closely related Cladocopium species. This highlights the power of aligning SymPortal outputs with the psbAncr marker for delineating Symbiodiniaceae species (Smith et al. 2020). Interestingly, across both A. cf. globiceps and A. curta, the C3f/C3fl-C3ae-C3-C3bj-C50a-C3h ITS2 profile completely dominated both microhabitats of a colony whenever it was present. This could be evidence of a species- or strain-level competitive hierarchy where certain taxa with wide photoacclimatory potentials outcompete other Symbiodiniaceae from all optical niches when they establish within a colony. Over long timescales, such competitive hierarchies could explain why some Symbiodiniaceae eventually reach near ubiquitous representation across many 100’s of kms of reef (Pettay and Lajeunesse 2013).
Hydnophora microconos as a new model species to study symbiont cohabitation
H. cf. microconos was particularly intriguing due to the inter- and intra-genus diversity of their dominant Symbiodiniaceae. Hydnophora spp. are known to acquire their symbionts horizontally (Fabina et al. 2012) and H. microconos has previously been reported to associate with either Cladocopium or Durusdinium on the same reef (LaJeunesse et al. 2003). We propose that H. microconos may serve as an ideal model species for further investigations of symbiont cohabitation and may be considered as a Pacific analogue to the model coral species Orbicella faveolata in the Caribbean (Kemp et al. 2014). Inherent symbiont flexibility may enhance the potential for colonisation of a cohort of symbiont species [potentially during the recruitment phase (Quigley et al. 2017)] that subsequently become partitioned across multiple optical niches as the colony grows. Indeed, symbiont flexibility may be integral for allowing symbiont species to occupy distinct optical niches in perpetuity.
In H. cf. microconos, the extent of phylogenetic distance among low-abundance sequences across shaded and exposed fragments within a single colony was strongly positively correlated with the magnitude of difference in nonphotochemical quenching utilisation [1 – Q]. The capacity to preferentially dissipate excitation energy via [1 – Q] is an important trait for tolerating light stress and is known to broadly correlate with Symbiodiniaceae genera (Robison and Warner 2006; Suggett et al. 2015). However, preferential nonphotochemical quenching reliance can also vary between isolates of the same species, highlighting the capacity of Symbiodiniaceae to adapt to differing environmental histories (Howells et al. 2012; Suggett et al. 2015). Differences in rare symbiont diversity between microhabitats may be partially driven by the selection for symbionts with a high capacity for nonphotochemical quenching in exposed colony regions. However, rare symbionts likely do not contribute strongly to the fluorescence signal of the PAM and, therefore, it is not possible to discern if these rare symbionts share this phenotype. The extent of nonphotochemical quenching in dominant symbiont taxa may serve as diagnostic of distinct optical niches and a proxy for zones likely harbouring diverse background symbiont species.
Conclusion
Due to the rapid nature of climate change, coral reef resilience may largely depend on capacity for adaptive shifting of pre-existing symbiont diversity within each colony (Cunning et al. 2015; Silverstein et al. 2015). Predicting rearrangements in symbiont species may require a theoretical framework that incorporates differential symbiont performance, competition, changing niche spaces, and successional processes within the holobiont, i.e. an ecology of Symbiodiniaceae interactions within corals. Gradients of irradiance across coral colonies may partially explain the coexistence of diverse symbiont taxa, and our data suggest that pooling sequences from exposed and shaded niches of individual colonies is an effective method for capturing the extent of within-colony Symbiodiniaceae diversity. However, future studies are needed to answer whether sampling colony extremities are sufficient to capture total within-colony Symbiodiniaceae diversity, or whether multi-variable niches (e.g. light × water flow/nutrient exchange combinations) promote additional diversity. Niche-partitioning may enhance the genomic variation available within the host, strengthening the ability of corals to maintain their adaptive potential. Validating the light-driven niche-differentiation hypothesis will ultimately require more observational studies and experimental manipulation of coral colonies. As corals face increasingly threatening circumstances globally, symbiont niche-differentiation should promptly inspire further investigation.
References
Abrego D, Van Oppen MJH, Willis BL (2009) Onset of algal endosymbiont specificity varies among closely related species of Acropora corals during early ontogeny. Mol Ecol 18:3532–3543. https://doi.org/10.1111/j.1365-294X.2009.04276.x
Abrego D, Ulstrup KE, Willis BL, van Oppen MJH (2008) Species-specific interactions between algal endosymbionts and coral hosts define their bleaching response to heat and light stress. Proc Royal Soc B 275:2273–2282. https://doi.org/10.1098/rspb.2008.0180
Anthony KR, Hoegh-Guldberg O (2003) Kinetics of photoacclimation in corals. Oecologia 134:23–31. https://doi.org/10.1007/s00442-002-1095-1
Anthony KRN, Hoogenboom MO, Connolly SR (2005) Adaptive variation in coral geometry and the optimization of internal colony light climates. Funct Ecol 19:17–26. https://doi.org/10.1111/j.0269-8463.2005.00925.x
Arif C, Daniels C, Bayer T, Banguera-Hinestroza E, Barbrook A, Howe CJ, LaJeunesse TC, Voolstra CR (2014) Assessing Symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Mol Ecol 23:4418–4433. https://doi.org/10.1111/mec.12869
Baker AC, Starger CJ, McClanahan TR, Glynn PW (2004) Corals’ adaptive response to climate change. Nature 430:741–741. https://doi.org/10.1038/430741a
Bay LK, Doyle J, Logan M, Berkelmans R (2016) Recovery from bleaching is mediated by threshold densities of background thermo-tolerant symbiont types in a reef-building coral. R Soc Open Sci 3:160322. https://doi.org/10.1098/rsos.160322
Berkelmans R, Van Oppen MJ (2006) The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’for coral reefs in an era of climate change. Proc Royal Soc B 273:2305–2312. https://doi.org/10.1098/rspb.2006.3567
Boulotte NM, Dalton SJ, Carroll AG, Harrison PL, Putnam HM, Peplow LM, van Oppen MJH (2016) Exploring the Symbiodinium rare biosphere provides evidence for symbiont switching in reef-building corals. ISME J 10:2693–2701. https://doi.org/10.1038/ismej.2016.54
Brown B, Downs C, Dunne R, Gibb S (2002) Exploring the basis of thermotolerance in the reef coral Goniastrea aspera. Mar Ecol-Prog Ser 242:119–129. https://doi.org/10.3354/meps242119
Brown BE, Dunne RP, Goodson MS, Douglas AE (2000a) Bleaching patterns in reef corals. Nature 404:142–143. https://doi.org/10.1038/35004657
Brown BE, Dunne RP, Warner ME, Ambarsari I, Fitt WK, Gibb SW, Cummings DG (2000b) Damage and recovery of Photosystem II during a manipulative field experiment on solar bleaching in the coral Goniastrea aspera. Mar Ecol-Prog Ser 195:117–124. https://doi.org/10.3354/meps195117
Buddemeier RW, Fautin DG (1993) Coral bleaching as an adaptive mechanism: a testable hypothesis. Bioscience 43:320–326. https://doi.org/10.2307/1312064
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581. https://doi.org/10.1038/nmeth.3869
Celliers L, Schleyer MH (2002) Coral bleaching on high-latitude marginal reefs at Sodwana Bay, South Africa. Mar Pollut Bull 44:1380–1387. https://doi.org/10.1016/S0025-326X(02)00302-8
Claar DC, Starko S, Tietjen KL, Epstein HE, Cunning R, Cobb KM, Baker AC, Gates RD, Baum JK (2020) Dynamic symbioses reveal pathways to coral survival through prolonged heatwaves. Nat Commun 11:6097. https://doi.org/10.1038/s41467-020-19169-y
Cohen I, Dubinsky Z (2015) Long term photoacclimation responses of the coral Stylophora pistillata to reciprocal deep to shallow transplantation: photosynthesis and calcification. Front Mar Sci 2:45. https://doi.org/10.3389/fmars.2015.00045
Cowman PF, Quattrini AM, Bridge TCL, Watkins-Colwell GJ, Fadli N, Grinblat M, Roberts TE, McFadden CS, Miller DJ, Baird AH (2020) An enhanced target-enrichment bait set for Hexacorallia provides phylogenomic resolution of the staghorn corals (Acroporidae) and close relatives. Mol Phylogenet Evol 153:106944. https://doi.org/10.1016/j.ympev.2020.106944
Cunning R, Silverstein RN, Baker AC (2015) Investigating the causes and consequences of symbiont shuffling in a multi-partner reef coral symbiosis under environmental change. Proc Royal Soc B 282:20141725. https://doi.org/10.1098/rspb.2014.1725
Cunning R, Ritson-Williams R, Gates RD (2016) Patterns of bleaching and recovery of Montipora capitata in Kāne’ohe Bay, Hawai’i, USA. Mar Ecol-Prog Ser 551:131–139. https://doi.org/10.3354/meps11733
Cunning R, Silverstein RN, Baker AC (2018) Symbiont shuffling linked to differential photochemical dynamics of Symbiodinium in three Caribbean reef corals. Coral Reefs 37:145–152. https://doi.org/10.1007/s00338-017-1640-3
D’Angelo C, Denzel A, Vogt A, Matz MV, Oswald F, Salih A, Nienhaus GU, Wiedenmann J (2008) Blue light regulation of host pigment in reef-building corals. Mar Ecol-Prog Ser 364:97–106. https://doi.org/10.3354/meps07588
Davy SK, Allemand D, Weis VM (2012) Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev 76:229. https://doi.org/10.1128/MMBR.05014-11
Díaz-Almeyda EM, Prada C, Ohdera AH, Moran H, Civitello DJ, Iglesias-Prieto R, Carlo TA, LaJeunesse TC, Medina M (2017) Intraspecific and interspecific variation in thermotolerance and photoacclimation in Symbiodinium dinoflagellates. Proc Royal Soc B 284:20171767. https://doi.org/10.1098/rspb.2017.1767
Dixon P (2003) VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. https://doi.org/10.1111/j.1654-1103.2003.tb02228.x
Enríquez S, Méndez ER, Hoegh-Guldberg O, Iglesias-Prieto R (2017) Key functional role of the optical properties of coral skeletons in coral ecology and evolution. Proc Royal Soc B 284:20161667. https://doi.org/10.1098/rspb.2016.1667
Fabina NS, Putnam HM, Franklin EC, Stat M, Gates RD (2012) Transmission mode predicts specificity and interaction patterns in coral-Symbiodinium networks. PLoS ONE 7:e44970. https://doi.org/10.1371/journal.pone.0044970
Frade PR, De Jongh F, Vermeulen F, Van Bleijswijk J, Bak RPM (2008) Variation in symbiont distribution between closely related coral species over large depth ranges. Mol Ecol 17:691–703. https://doi.org/10.1111/j.1365-294X.2007.03612.x
Gabay Y, Weis VM, Davy SK (2018) Symbiont identity influences patterns of symbiosis establishment, host growth, and asexual reproduction in a model cnidarian-dinoflagellate symbiosis. Biol Bull 234:1–10. https://doi.org/10.1086/696365
Gabay Y, Parkinson JE, Wilkinson SP, Weis VM, Davy SK (2019) Inter-partner specificity limits the acquisition of thermotolerant symbionts in a model cnidarian-dinoflagellate symbiosis. ISME J 13:2489–2499. https://doi.org/10.1038/s41396-019-0429-5
Garren M, Walsh S, Caccone A, Knowlton N (2006) Patterns of association between Symbiodinium and members of the Montastraea annularis species complex on spatial scales ranging from within colonies to between geographic regions. Coral Reefs 25:503–512. https://doi.org/10.1007/s00338-006-0146-1
Gattuso J-P, Allemand D, Frankignoulle M (1999) Photosynthesis and calcification at cellular, organismal and community levels in coral reefs: a review on interactions and control by carbonate chemistry. Am Zool 39:160–183. https://doi.org/10.1093/icb/39.1.160
Goulet TL, Lucas MQ, Schizas NV (2019) Symbiodiniaceae genetic diversity and symbioses with hosts from shallow to mesophotic coral ecosystems. In: Loya Y, Puglise KA, Bridge TCL (eds) Mesophotic coral ecosystems. Springer International Publishing, Cham, pp 537–551
Hennige S, Suggett D, Warner M, McDougall K, Smith D (2009) Photobiology of Symbiodinium revisited: bio-physical and bio-optical signatures. Coral Reefs 28:179–195. https://doi.org/10.1007/s00338-008-0444-x
Hennige SJ, Smith DJ, Perkins R, Consalvey M, Paterson DM, Suggett DJ (2008) Photoacclimation, growth and distribution of massive coral species in clear and turbid waters. Mar Ecol-Prog Ser 369:77–88. https://doi.org/10.3354/meps07612
Hill R, Ralph PJ (2005) Diel and seasonal changes in fluorescence rise kinetics of three scleractinian corals. Funct Plant Biol 32:549–559. https://doi.org/10.1071/FP05017
Hoadley KD, Pettay DT, Lewis A, Wham D, Grasso C, Smith R, Kemp DW, Lajeunesse T, Warner ME (2021) Different functional traits among closely related algal symbionts dictate stress endurance for vital Indo-Pacific reef-building corals. Glob Chang Biol 27:5295–5309. https://doi.org/10.1111/gcb.15799
Hoeksema BW, Cairns S (2021) World List of Scleractinia. Accessed at http://www.marinespecies.org/scleractinia on 2021-12-12
Howells EJ, Beltran VH, Larsen NW, Bay LK, Willis BL, van Oppen MJH (2012) Coral thermal tolerance shaped by local adaptation of photosymbionts. Nat Clim Chang 2:116. https://doi.org/10.1038/nclimate1330
Hume BC, Mejia-Restrepo A, Voolstra CR, Berumen ML (2020) Fine-scale delineation of Symbiodiniaceae genotypes on a previously bleached central Red Sea reef system demonstrates a prevalence of coral host-specific associations. Coral Reefs. https://doi.org/10.1007/s00338-020-01917-7
Hume BCC, Smith EG, Ziegler M, Warrington HJM, Burt JA, LaJeunesse TC, Wiedenmann J, Voolstra CR (2019) SymPortal: a novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol Ecol Resour 19:1063–1080. https://doi.org/10.1111/1755-0998.13004
Iglesias-Prieto R, Trench RK (1994) Acclimation and adaptation to irradiance in symbiotic dinoflagellates. I. Responses of the photosynthetic unit to changes in photon flux density. Mar Ecol-Prog Ser 113:163–175. https://doi.org/10.3354/meps113163
Iglesias-Prieto R, Trench RK (1997) Acclimation and adaptation to irradiance in symbiotic dinoflagellates. II. Response of chlorophyll–protein complexes to different photon-flux densities. Mar Biol 130:23–33. https://doi.org/10.1007/s002270050221
Kaniewska P, Anthony KRN, Hoegh-Guldberg O (2008) Variation in colony geometry modulates internal light levels in branching corals, Acropora humilis and Stylophora pistillata. Mar Biol 155:649–660. https://doi.org/10.1007/s00227-008-1061-5
Kemp DW, Fitt WK, Schmidt GW (2008) A microsampling method for genotyping coral symbionts. Coral Reefs 27:289–293. https://doi.org/10.1007/s00338-007-0333-8
Kemp DW, Hernandez-Pech X, Iglesias-Prieto R, Fitt WK, Schmidt GW (2014) Community dynamics and physiology of Symbiodinium spp. before, during, and after a coral bleaching event. Limnol Oceanogr 59:788–797. https://doi.org/10.4319/lo.2014.59.3.0788
LaJeunesse TC, Thornhill DJ (2011) Improved resolution of reef-coral endosymbiont (Symbiodinium) species diversity, ecology, and evolution through psba non-coding region genotyping. PLoS ONE 6:e29013. https://doi.org/10.1371/journal.pone.0029013
LaJeunesse TC, Smith RT, Finney J, Oxenford H (2009) Outbreak and persistence of opportunistic symbiotic dinoflagellates during the 2005 Caribbean mass coral ‘bleaching’event. Proc Royal Soc B 276:4139–4148. https://doi.org/10.1098/rspb.2009.1405
LaJeunesse TC, Loh WK, Van Woesik R, Hoegh-Guldberg O, Schmidt GW, Fitt WK (2003) Low symbiont diversity in southern Great Barrier Reef corals, relative to those of the Caribbean. Limnol Oceanogr 48:2046–2054. https://doi.org/10.4319/lo.2003.48.5.2046
LaJeunesse TC, Parkinson JE, Gabrielson PW, Jeong HJ, Reimer JD, Voolstra CR, Santos SR (2018) Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr Biol 28:2570-2580.e2576. https://doi.org/10.1016/j.cub.2018.07.008
LaJeunesse TC, Pettay DT, Sampayo EM, Phongsuwan N, Brown B, Obura DO, Hoegh-Guldberg O, Fitt WK (2010) Long-standing environmental conditions, geographic isolation and host–symbiont specificity influence the relative ecological dominance and genetic diversification of coral endosymbionts in the genus Symbiodinium. J Biogeogr 37:785–800. https://doi.org/10.1111/j.1365-2699.2010.02273.x
Lee MJ, Jeong HJ, Jang SH, Lee SY, Kang NS, Lee KH, Kim HS, Wham DC, LaJeunesse TC (2016) Most low-abundance “background” Symbiodinium spp. are transitory and have minimal functional significance for symbiotic corals. Microb Ecol 71:771–783. https://doi.org/10.1007/s00248-015-0724-2
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228. https://doi.org/10.1128/AEM.71.12.8228-8235.2005
MacIntyre HL, Kana TM, Anning T, Geider RJ (2002) Photoacclimation of photosynthesis irradiance response curves and photosynthetic pigments in microalgae and cyanobacteria. J Phycol 38:17–38. https://doi.org/10.1046/j.1529-8817.2002.00094.x
Maia R, Eliason CM, Bitton PP, Doucet SM, Shawkey MD (2013) pavo: an R package for the analysis, visualization and organization of spectral data. Methods Ecol Evol 4:906–913. https://doi.org/10.1111/2041-210X.12069
Marcelino LA, Westneat MW, Stoyneva V, Henss J, Rogers JD, Radosevich A, Turzhitsky V, Siple M, Fang A, Swain TD, Fung J, Backman V (2013) Modulation of Light-Enhancement to Symbiotic Algae by Light-Scattering in Corals and Evolutionary Trends in Bleaching. PLoS ONE 8:e61492. https://doi.org/10.1371/journal.pone.0061492
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17:3. https://doi.org/10.14806/ej.17.1.200
Matthews JL, Crowder CM, Oakley CA, Lutz A, Roessner U, Meyer E, Grossman AR, Weis VM, Davy SK (2017) Optimal nutrient exchange and immune responses operate in partner specificity in the cnidarian-dinoflagellate symbiosis. Proc Natl Acad Sci 114:13194. https://doi.org/10.1073/pnas.1710733114
McGinley MP, Aschaffenburg MD, Pettay DT, Smith RT, LaJeunesse TC, Warner ME (2012) Symbiodinium spp. in colonies of eastern Pacific Pocillopora spp. are highly stable despite the prevalence of low-abundance background populations. Mar Ecol-Prog Ser 462:1–7. https://doi.org/10.3354/meps09914
McIlroy SE, Wong JCY, Baker DM (2020) Competitive traits of coral symbionts may alter the structure and function of the microbiome. ISME J 14:2424–2432. https://doi.org/10.1038/s41396-020-0697-0
McIlroy SE, Cunning R, Baker AC, Coffroth MA (2019) Competition and succession among coral endosymbionts. Ecol Evol 9:12767–12778. https://doi.org/10.1002/ece3.5749
McMurdie PJ, Holmes S (2013) Phyloseq: an r package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. https://doi.org/10.1371/journal.pone.0061217
Mieog JC, van Oppen MJ, Cantin NE, Stam WT, Olsen JL (2007) Real-time PCR reveals a high incidence of Symbiodinium clade D at low levels in four scleractinian corals across the Great Barrier Reef: implications for symbiont shuffling. Coral Reefs 26:449–457. https://doi.org/10.1007/s00338-007-0244-8
Miller MA, Pfeiffer W, Schwartz T (2011) The CIPRES science gateway: a community resource for phylogenetic analyses. Proceedings of the 2011 TeraGrid Conference: extreme digital discovery, pp. 1–8
Muscatine L (1990) The role of symbiotic algae in carbon and energy flux in reef corals. Coral Reefs 25:1–29
Nitschke MR, Gardner SG, Goyen S, Fujise L, Camp EF, Ralph PJ, Suggett DJ (2018) Utility of photochemical traits as diagnostics of thermal tolerance amongst great barrier reef corals. Front Mar Sci 5:45. https://doi.org/10.3389/fmars.2018.00045
Nitschke MR, Craveiro SC, Brandão C, Fidalgo C, Serôdio J, Calado AJ, Frommlet JC (2020) Description of Freudenthalidium gen. nov. and Halluxium gen. nov. to formally recognize clades Fr3 and H as genera in the family Symbiodiniaceae (Dinophyceae). J Phycol 56:923–940. https://doi.org/10.1111/jpy.12999
Oladi M, Rouzbehani S, Ahmadzadeh F, Ghazilou A (2021) Dynamics of Dipsastraea pallida-symbiont association following bleaching events across the northern Persian Gulf and Gulf of Oman. Symbiosis 84:141–149. https://doi.org/10.1007/s13199-021-00773-5
Oliver TA, Palumbi SR (2011) Many corals host thermally resistant symbionts in high-temperature habitat. Coral Reefs 30:241–250. https://doi.org/10.1007/s00338-010-0696-0
Ow Y, Todd P (2010) Light-induced morphological plasticity in the scleractinian coral Goniastrea pectinata and its functional significance. Coral Reefs 29:797–808. https://doi.org/10.1007/s00338-010-0631-4
Penin L, Adjeroud M, Schrimm M, Lenihan HS (2007) High spatial variability in coral bleaching around Moorea (French Polynesia): patterns across locations and water depths. C R Biol 330:171–181. https://doi.org/10.1016/j.crvi.2006.12.003
Pettay DT, Lajeunesse TC (2013) Long-range dispersal and high-latitude environments influence the population structure of a “stress-tolerant” dinoflagellate endosymbiont. PLoS ONE 8:e79208–e79208. https://doi.org/10.1371/journal.pone.0079208
Pettay DT, Wham DC, Smith RT, Iglesias-Prieto R, LaJeunesse TC (2015) Microbial invasion of the Caribbean by an Indo-Pacific coral zooxanthella. Proc Natl Acad Sci 112:7513. https://doi.org/10.1073/pnas.1502283112
Pochon X, LaJeunesse TC (2021) Miliolidium n. gen, a new Symbiodiniacean genus whose members associate with soritid foraminifera or are free-living. J Eukaryot Microbiol 68:e12856. https://doi.org/10.1111/jeu.12856
Pochon X, Pawlowski J, Zaninetti L, Rowan R (2001) High genetic diversity and relative specificity among Symbiodinium-like endosymbiotic dinoflagellates in soritid foraminiferans. Mar Biol 139:1069–1078. https://doi.org/10.1007/s002270100674
Quigley KM, Willis BL, Bay LK (2017) Heritability of the Symbiodinium community in vertically- and horizontally-transmitting broadcast spawning corals. Sci Rep 7:8219. https://doi.org/10.1038/s41598-017-08179-4
Quigley KM, Davies SW, Kenkel CD, Willis BL, Matz MV, Bay LK (2014) Deep-sequencing method for quantifying background abundances of Symbiodinium types: exploring the rare Symbiodinium biosphere in reef-building corals. PLoS ONE. https://doi.org/10.1371/journal.pone.0094297
Reynolds JM, Bruns BU, Fitt WK, Schmidt GW (2008) Enhanced photoprotection pathways in symbiotic dinoflagellates of shallow-water corals and other cnidarians. Proc Natl Acad Sci 105:13674. https://doi.org/10.1073/pnas.0805187105
Robison JD, Warner ME (2006) Differential impacts of photoacclimation and thermal stress on the photobiology of four different phylotypes of Symbiodinium (Pyrrhophyta). J Phycol 42:568–579. https://doi.org/10.1111/j.1529-8817.2006.00232.x
Rowan R (2004) Thermal adaptation in reef coral symbionts. Nature 430:742–742. https://doi.org/10.1038/430742a
Rowan R, Knowlton N (1995) Intraspecific diversity and ecological zonation in coral-algal symbiosis. Proc Natl Acad Sci 92:2850. https://doi.org/10.1073/pnas.92.7.2850
Rowan R, Knowlton N, Baker A, Jara J (1997) Landscape ecology of algal symbionts creates variation in episodes of coral bleaching. Nature 388:265–269. https://doi.org/10.1038/40843
Sampayo EM, Franceschinis L, Hoegh-Guldberg O, Dove S (2007) Niche partitioning of closely related symbiotic dinoflagellates. Mol Ecol 16:3721–3733. https://doi.org/10.1111/j.1365-294X.2007.03403.x
Sampayo EM, Ridgway T, Bongaerts P, Hoegh-Guldberg O (2008) Bleaching susceptibility and mortality of corals are determined by fine-scale differences in symbiont type. Proc Natl Acad Sci 105:10444. https://doi.org/10.1073/pnas.0708049105
Seutin G, White BN, Boag PT (1991) Preservation of avian blood and tissue samples for DNA analyses. Can J Zool 69:82–90. https://doi.org/10.1139/z91-013
Silverstein RN, Correa AM, Baker AC (2012) Specificity is rarely absolute in coral–algal symbiosis: implications for coral response to climate change. Proc Royal Soc B 279:2609–2618. https://doi.org/10.1098/rspb.2012.0055
Silverstein RN, Cunning R, Baker AC (2015) Change in algal symbiont communities after bleaching, not prior heat exposure, increases heat tolerance of reef corals. Glob Chang Biol 21:236–249. https://doi.org/10.1111/gcb.12706
Smith EG, Gurskaya A, Hume BCC, Voolstra CR, Todd PA, Bauman AG, Burt JA (2020) Low Symbiodiniaceae diversity in a turbid marginal reef environment. Coral Reefs 39:545–553. https://doi.org/10.1007/s00338-020-01956-0
Sproles AE, Oakley CA, Krueger T, Grossman AR, Weis VM, Meibom A, Davy SK (2020) Sub-cellular imaging shows reduced photosynthetic carbon and increased nitrogen assimilation by the non-native endosymbiont Durusdinium trenchii in the model cnidarian Aiptasia. Environ Microbiol 22:3741–3753. https://doi.org/10.1111/1462-2920.15142
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Stat M, Gates RD (2011) Clade D Symbiodinium in scleractinian corals: a “nugget” of hope, a selfish opportunist, an ominous sign, or all of the above? J Mar Biol. https://doi.org/10.1155/2011/730715
Stat M, Pochon X, Cowie ROM, Gates RD (2009a) Specificity in communities of Symbiodinium in corals from Johnston Atoll. Mar Ecol Prog Ser 386:83–96. https://doi.org/10.3354/meps08080
Stat M, Loh WKW, LaJeunesse TC, Hoegh-Guldberg O, Carter DA (2009b) Stability of coral–endosymbiont associations during and after a thermal stress event in the southern Great Barrier Reef. Coral Reefs 28:709–713. https://doi.org/10.1007/s00338-009-0509-5
Suggett DJ, Warner ME, Leggat W (2017) Symbiotic dinoflagellate functional diversity mediates coral survival under ecological crisis. Trends Ecol Evol 32:735–745. https://doi.org/10.1016/j.tree.2017.07.013
Suggett DJ, Le Floc’H E, Harris GN, Leonardos N, Geider RJ (2007) Different strategies of photoacclimation by two strains of Emiliania huxleyi (Haptophyta) 1. J Phycol 43(6):1209–1222. https://doi.org/10.1111/j.1529-8817.2007.00406.x
Suggett DJ, Goyen S, Evenhuis C, Szabó M, Pettay DT, Warner ME, Ralph PJ (2015) Functional diversity of photobiological traits within the genus Symbiodinium appears to be governed by the interaction of cell size with cladal designation. New Phytol 208:370–381. https://doi.org/10.1111/nph.13483
Thomas L, Kendrick G, Kennington W, Richards Z, Stat M (2014) Exploring Symbiodinium diversity and host specificity in Acropora corals from geographical extremes of Western a ustralia with 454 amplicon pyrosequencing. Mol Ecol 23:3113–3126. https://doi.org/10.1111/mec.12801
Thornhill DJ, Lewis AM, Wham DC, LaJeunesse TC (2014) Host-specialist lineages dominate the adaptive radiation of reef coral endosymbionts. Evolution 68:352–367. https://doi.org/10.1111/evo.12270
Thornhill DJ, LaJeunesse TC, Kemp DW, Fitt WK, Schmidt GW (2006) Multi-year, seasonal genotypic surveys of coral-algal symbioses reveal prevalent stability or post-bleaching reversion. Mar Biol 148:711–722. https://doi.org/10.1007/s00227-005-0114-2
van Oppen MJH, Palstra FP, Piquet AMT, Miller DJ (2001) Patterns of coral–dinoflagellate associations in Acropora: significance of local availability and physiology of Symbiodinium strains and host–symbiont selectivity. Proc Royal Soc B 268:1759–1767. https://doi.org/10.1098/rspb.2001.1733
Veron J (2000) Corals of the World. Australian Institute of Marine Science, Townsville
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Warner ME, Lesser MP, Ralph PJ (2010) Chlorophyll fluorescence in reef building corals. In: Suggett DJ, Prášil O, Borowitzka MA (eds) Chlorophyll a fluorescence in aquatic sciences: methods and applications. Springer, Netherlands, Dordrecht, pp 209–222
Wilkinson S (2018) kmer: an R package for fast alignment-free clustering of biological sequences
Winters G, Beer S, Zvi BB, Brickner I, Loya Y (2009) Spatial and temporal photoacclimation of Stylophora pistillata: zooxanthella size, pigmentation, location and clade. Mar Ecol-Prog Ser 384:107–119. https://doi.org/10.3354/meps08036
Wright ES (2016) Using DECIPHER v2. 0 to analyze big biological sequence data in R. R Journal 8:352–359
Yokouchi H, Takeyama H, Miyashita H, Maruyama T, Matsunaga T (2003) In situ identification of symbiotic dinoflagellates, the genus Symbiodinium with fluorescence-labeled rRNA-targeted oligonucleotide probes. J Microbiol Methods 53:327–334. https://doi.org/10.1016/s0167-7012(02)00250-6
Yorifuji M, Yamashita H, Suzuki G, Kawasaki T, Tsukamoto T, Okada W, Tamura K, Nakamura R, Inoue M, Yamazaki M, Harii S (2021) Unique environmental Symbiodiniaceae diversity at an isolated island in the northwestern Pacific. Mol Phylogenet Evol 161:107158. https://doi.org/10.1016/j.ympev.2021.107158
Ziegler M, Eguíluz VM, Duarte CM, Voolstra CR (2018) Rare symbionts may contribute to the resilience of coral–algal assemblages. ISME J 12:161–172. https://doi.org/10.1038/ismej.2017.151
Acknowledgements
MRN was supported by a Rutherford Foundation Postdoctoral Fellowship (Royal Society Te Apārangi) and a National Geographic Explorers Grant (EC-66454R-20). REL was supported by the Alison Morton Scholarship in Ecology/Marine Biology, the Wellington Master’s by Thesis scholarship, and research funding from the Wellington Faculty of Science (to SKD). All collections were performed by MRN, SGG and TR under Research Permit #408435 (Office of the Prime Minister, Cook Islands) awarded to MRN and TR. We acknowledge the expertise and guidance of Tom Bridge in assigning taxonomic identifiers to collected specimens.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lewis, R.E., Davy, S.K., Gardner, S.G. et al. Colony self-shading facilitates Symbiodiniaceae cohabitation in a South Pacific coral community. Coral Reefs 41, 1433–1447 (2022). https://doi.org/10.1007/s00338-022-02292-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-022-02292-1