Abstract
Key message
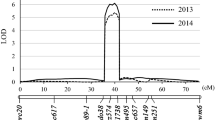
Six major QTLs for wheat grain size and weight were identified on chromosomes 4A, 4B, 5A and 6A across multiple environments, and were validated in different genetic backgrounds.
Abstract
Grain size and weight are crucial components of wheat yield. Dissection of their genetic control is thus essential for the improvement of yield potential in wheat breeding. We used a doubled haploid (DH) population to detect quantitative trait loci (QTLs) for grain width (GW), grain length (GL), and thousand grain weight (TGW) in five environments. Six major QTLs, QGw.cib-4B.2, QGl.cib-4A, QGl.cib-5A.1, QGl.cib-6A, QTgw.cib-4B, and QTgw.cib-5A, were consistently identified in at least three individual environments and in best linear unbiased prediction (BLUP) datasets, and explained 5.65–34.06% of phenotypic variation. QGw.cib-4B.2, QTgw.cib-4B, QGl.cib-5A.1 and QGl.cib-6A had no effect on grain number per spike (GNS). In addition to QGl.cib-4A, the other major QTLs were further validated by using Kompetitive Allele Specific PCR (KASP) markers in different genetic backgrounds. Moreover, significant interactions between the three major GL QTLs and two major TGW QTLs were observed. Comparison analysis showed that QGl.cib-5A.1 and QGl.cib-6A are likely new loci. Notably, QGw.cib-4B.2 and QTgw.cib-4B were co-located on chromosome 4B and improved TGW by increasing only GW, unlike nearby or overlapped loci reported previously. Three genes associated with grain development within the QGw.cib-4B.2/QTgw.cib-4B interval were identified by searches on sequence similarity, spatial expression patterns, and orthologs. The major QTLs and KASP markers reported here will be useful for elucidating the genetic architecture of grain size and weight and for developing new wheat cultivars with high and stable yield.








Similar content being viewed by others
References
Ashikari M, Wu J, Yano M, Sasaki T, Yoshimura A (1999) Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proc Natl Acad Sci U S A 96:10284–10289. https://doi.org/10.1073/pnas.96.18.10284
Borrill P, Ramirez-Gonzalez R, Uauy C (2016) expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol 170:2172–2186. https://doi.org/10.1104/pp.15.01667
Butterworth MH, Semenov MA, Barnes A, Moran D, West JS, Fitt BDL (2010) North-South divide: contrasting impacts of climate change on crop yields in Scotland and England. J R Soc Interface 7:123–130. https://doi.org/10.1098/rsif.2009.0111
Cao P, Liang X, Zhao H, Feng B, Xu E, Wang L et al (2019) Identification of the quantitative trait loci controlling spike-related traits in hexaploid wheat (Triticum aestivum L.). Planta 250:1967–1981. https://doi.org/10.1007/s00425-019-03278-0
Cao S, Xu D, Hanif M, Xia X, He Z (2020) Genetic architecture underpinning yield component traits in wheat. Theor Appl Genet 133:1811–1823. https://doi.org/10.1007/s00122-020-03562-8
Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW et al (2011) An atypical heterotrimeric G-protein β-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J 67:840–851. https://doi.org/10.1111/j.1365-313X.2011.04638.x
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y et al (2020a) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Chen Y, Song W, Xie X, Wang Z, Guan P, Peng H et al (2020b) A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the triticeae tribe as a pilot practice in the plant pangenomic era. Mol Plant 13:1694–1708. https://doi.org/10.1016/j.molp.2020.09.019
Chen Y, Yan Y, Wu TT, Zhang GL, Yin H, Chen W et al (2020c) Cloning of wheat keto-acyl thiolase 2B reveals a role of jasmonic acid in grain weight determination. Nat Commun. https://doi.org/10.1038/s41467-020-20133-z
Cheng X, Xin M, Xu R, Chen Z, Cai W, Chai L et al (2020) A single amino acid substitution in STKc_GSK3 kinase conferring semispherical grains and its implications for the origin of triticum sphaerococcum. Plant Cell 32:923–934. https://doi.org/10.1105/TPC.19.00580
Cheng ZJ, Zhao XY, Shao XX, Wang F, Zhou C, Liu YG et al (2014) Abscisic acid regulates early seed development in arabidopsis by ABI5-Mediated transcription of SHORT HYPOCOTYL UNDER BLUE1. Plant Cell 26:1053–1068. https://doi.org/10.1105/tpc.113.121566
Cui F, Zhao C, Ding A, Li J, Wang L, Li X et al (2014) Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor Appl Genet 127:659–675. https://doi.org/10.1007/s00122-013-2249-8
Cui Y, Zhu M, Xu Z, Xu Q (2019) Assessment of the effect of ten heading time genes on reproductive transition and yield components in rice using a CRISPR/Cas9 system. Theor Appl Genet 132:1887–1896. https://doi.org/10.1007/s00122-019-03324-1
Dholakia BB, Ammiraju JSS, Singh H, Lagu MD, Röder MS, Rao VS et al (2003) Molecular marker analysis of kernel size and shape in bread wheat. Plant Breed 122:392–395. https://doi.org/10.1046/j.1439-0523.2003.00896.x
Dong L, Wang F, Liu T, Dong Z, Li A, Jing R et al (2014) Natural variation of TaGASR7-A1 affects grain length in common wheat under multiple cultivation conditions. Mol Breed 34:937–947. https://doi.org/10.1007/s11032-014-0087-2
Duan P, Rao Y, Zeng D, Yang Y, Xu R, Zhang B et al (2014) SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J 77:547–557. https://doi.org/10.1111/tpj.12405
Evans N, Baierl A, Semenov MA, Gladders P, Fitt BDL (2008) Range and severity of a plant disease increased by global warming. J R Soc Interface 5:525–531. https://doi.org/10.1098/rsif.2007.1136
Fan X, Cui F, Zhao C, Zhang W, Yang L, Zhao X et al (2015) QTLs for flag leaf size and their influence on yield-related traits in wheat (Triticum aestivum L.). Mol Breed. https://doi.org/10.1007/s11032-015-0205-9
Fang N, Xu R, Huang L, Zhang B, Duan P, Li N et al (2016) SMALL GRAIN 11 controls grain size, grain number and grain yield in rice. Rice. https://doi.org/10.1186/s12284-016-0136-z
Fujisawa Y, Kato T, Ohki S, Ishikawa A, Kitano H, Sasaki T et al (1999) Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc Natl Acad Sci U S A 96:7575–7580. https://doi.org/10.1073/pnas.96.13.7575
Gegas VC, Nazari A, Griffiths S, Simmonds J, Fish L, Orford S et al (2010) A genetic framework for grain size and shape variation in wheat. Plant Cell 22:1046–1056. https://doi.org/10.1105/tpc.110.074153
Geng J, Li L, Lv Q, Zhao Y, Liu Y, Zhang L et al (2017) TaGW2-6A allelic variation contributes to grain size possibly by regulating the expression of cytokinins and starch-related genes in wheat. Planta 246:1153–1163. https://doi.org/10.1007/s00425-017-2759-8
Guan P, Di N, Mu Q, Shen X, Wang Y, Wang X et al (2019) Use of near-isogenic lines to precisely map and validate a major QTL for grain weight on chromosome 4AL in bread wheat (Triticum aestivum L.). Theor Appl Genet 132:2367–2379. https://doi.org/10.1007/s00122-019-03359-4
Guan P, Lu L, Jia L, Kabir MR, Zhang J, Lan T et al (2018) Global QTL analysis identifies genomic regions on chromosomes 4A and 4B harboring stable loci for yield-related traits across different environments in wheat (Triticum aestivum L.). Front Plant Sci. https://doi.org/10.3389/fpls.2018.00529
Guan P, Shen X, Mu Q, Wang Y, Wang X, Chen Y et al (2020) Dissection and validation of a QTL cluster linked to Rht-B1 locus controlling grain weight in common wheat (Triticum aestivum L.) using near-isogenic lines. Theor Appl Genet. https://doi.org/10.1007/s00122-020-03622-z
Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y et al (2005) The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17:2243–2254. https://doi.org/10.1105/tpc.105.030973
Hu J, Wang X, Zhang G, Jiang P, Chen W, Hao Y et al (2020) QTL mapping for yield-related traits in wheat based on four RIL populations. Theor Appl Genet. https://doi.org/10.1007/s00122-019-03515-w
Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H et al (2015) A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant 8:1455–1465. https://doi.org/10.1016/j.molp.2015.07.002
Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H et al (2013) Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat Genet 45:707–711. https://doi.org/10.1038/ng.2612
Jiang WB, Huang HY, Hu YW, Zhu SW, Wang ZY, Lin WH (2013) Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol 162:1965–1977. https://doi.org/10.1104/pp.113.217703
Khalid M, Afzal F, Gul A, Amir R, Subhani A, Ahmed Z et al (2019) Molecular characterization of 87 functional genes in wheat diversity panel and their association with phenotypes under well-watered and water-limited conditions. Front Plant Sci 10:1–15. https://doi.org/10.3389/fpls.2019.00717
Kumar A, Mantovani EE, Seetan R, Soltani A, Echeverry-Solarte M, Jain S et al (2016) Dissection of genetic factors underlying wheat Kernel shape and size in an Elite × Nonadapted cross using a high density SNP linkage map. Plant Genome. https://doi.org/10.3835/plantgenome2015.09.0081
Kumari S, Jaiswal V, Mishra VK, Paliwal R, Balyan HS, Gupta PK (2018) QTL mapping for some grain traits in bread wheat (Triticum aestivum L.). Physiol Mol Biol Plants 24:909–920. https://doi.org/10.1007/s12298-018-0552-1
Li F, Wen W, He Z, Liu J, Jin H, Cao S et al (2018) Genome - wide linkage mapping of yield - related traits in three Chinese bread wheat populations using high - density SNP markers. Theor Appl Genet 131:1903–1924. https://doi.org/10.1007/s00122-018-3122-6
Li F, Wen W, Liu J, Zhang Y, Cao S, He Z et al (2019a) Genetic architecture of grain yield in bread wheat based on genome-wide association studies. BMC Plant Biol 19:1–19. https://doi.org/10.1186/s12870-019-1781-3
Li N, Xu R, Li Y (2019b) Molecular networks of seed size control in plants. Annu Rev Plant Biol 70:435–463. https://doi.org/10.1146/annurev-arplant-050718-095851
Li Q, Li L, Yang X, Warburton ML, Bai G, Dai J et al (2010) Relationship, evolutionary fate and function of two maize co-orthologs of rice GW2 associated with kernel size and weight. BMC Plant Biol 10:1–15. https://doi.org/10.1186/1471-2229-10-143
Li T, Deng G, Su Y, Yang Z, Tang Y, Wang J et al (2021) Identification and validation of two major QTLs for spike compactness and length in bread wheat (Triticum aestivum L.) showing pleiotropic effects on yield-related traits. Theor Appl Genet. https://doi.org/10.1007/s00122-021-03918-8
Li T, Deng G, Tang Y, Su Y, Wang J, Cheng J (2021b) Identification and validation of a novel locus controlling spikelet number in bread wheat (Triticum aestivum L.). Front Plant Sci 12:1–14. https://doi.org/10.3389/fpls.2021.611106
Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L et al (2011) Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet 43:1266–1269. https://doi.org/10.1038/ng.977
Liu G, Jia L, Lu L, Qin D, Zhang J, Guan P et al (2014) Mapping QTLs of yield-related traits using RIL population derived from common wheat and Tibetan semi-wild wheat. Theor Appl Genet 127:2415–2432. https://doi.org/10.1007/s00122-014-2387-7
Liu T, Wu L, Gan X, Chen W, Liu B, Fedak G et al (2020) Mapping quantitative trait loci for 1000-grain weight in a double haploid population of common wheat. Int J Mol Sci. https://doi.org/10.3390/ijms21113960
Ma J, Zhang H, Li S, Zou Y, Li T, Liu J et al (2019) Identification of quantitative trait loci for kernel traits in a wheat cultivar Chuannong16. BMC Genet 20:1–12. https://doi.org/10.1186/s12863-019-0782-4
Mao H, Sun S, Yao J, Wang C, Yu S, Xu C et al (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci U S A 01:3–8. https://doi.org/10.1073/pnas.1014419107
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Mohler V, Albrecht T, Castell A, Diethelm M, Schweizer G, Hartl L (2016) Considering causal genes in the genetic dissection of kernel traits in common wheat. J Appl Genet 57:467–476. https://doi.org/10.1007/s13353-016-0349-2
Pang Y, Liu C, Wang D, St Amand P, Bernardo A, Li W et al (2020) High-resolution genome-wide association study identifies genomic regions and candidate genes for important agronomic traits in wheat. Mol Plant 13:1311–1327. https://doi.org/10.1016/j.molp.2020.07.008
Prashant R, Kadoo N, Desale C, Kore P, Dhaliwal HS, Chhuneja P et al (2012) Kernel morphometric traits in hexaploid wheat (Triticum aestivum L.) are modulated by intricate QTL × QTL and genotype × environment interactions. J Cereal Sci 56:432–439. https://doi.org/10.1016/j.jcs.2012.05.010
Ramírez-González RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L et al (2018) The transcriptional landscape of polyploid wheat. Science. https://doi.org/10.1126/science.aar6089
Ramya P, Chaubal A, Kulkarni K, Gupta L, Kadoo N, Dhaliwal HS et al (2010) QTL mapping of 1000-kernel weight, kernel length, and kernel width in bread wheat (Triticum aestivum L.). J Appl Genet 51:421–429. https://doi.org/10.1007/BF03208872
Segami S, Takehara K, Yamamoto T, Kido S, Kondo S, Iwasaki Y et al (2017) Overexpression of SRS5 improves grain size of brassinosteroid-related dwarf mutants in rice (Oryza sativa L.). Breed Sci 67:393–397. https://doi.org/10.1270/jsbbs.16198
Sestili F, Pagliarello R, Zega A, Saletti R, Pucci A, Botticella E et al (2019) Enhancing grain size in durum wheat using RNAi to knockdown GW2 genes. Theor Appl Genet 132:419–429. https://doi.org/10.1007/s00122-018-3229-9
Si L, Chen J, Huang X, Gong H, Luo J, Hou Q et al (2016) OsSPL13 controls grain size in cultivated rice. Nat Genet. https://doi.org/10.1038/ng.3518
Smith SE, Kuehl RO, Ray IM, Hui R, Soleri D (1998) Evaluation of simple methods for estimating broad-sense heritability in stands of randomly planted genotypes. Crop Sci 38:1125–1129. https://doi.org/10.2135/cropsci1998.0011183X003800050003x
Su Z, Jin S, Lu Y, Zhang G, Chao S, Bai G (2016) Single nucleotide polymorphism tightly linked to a major QTL on chromosome 7A for both kernel length and kernel weight in wheat. Mol Breed. https://doi.org/10.1007/s11032-016-0436-4
Sukumaran S, Lopes M, Dreisigacker S, Reynolds M (2018) Genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theor Appl Genet 131:985–998. https://doi.org/10.1007/s00122-017-3037-7
Sun XY, Wu K, Zhao Y, Kong FM, Han GZ, Jiang HM et al (2009) QTL analysis of kernel shape and weight using recombinant inbred lines in wheat. Euphytica 165:615–624. https://doi.org/10.1007/s10681-008-9794-2
Tu Y, Liu H, Liu J, Tang H, Mu Y, Deng M et al (2020) QTL mapping and validation of bread wheat flag leaf morphology across multiple environments in different genetic backgrounds. Theor Appl Genet. https://doi.org/10.1007/s00122-020-03695-w
Tura H, Edwards J, Gahlaut V, Garcia M, Sznajder B, Baumann U et al (2020) QTL analysis and fine mapping of a QTL for yield-related traits in wheat grown in dry and hot environments. Theor Appl Genet 133:239–257. https://doi.org/10.1007/s00122-019-03454-6
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X et al (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954. https://doi.org/10.1038/ng.2327
Wang X, Dong L, Hu J, Pang Y, Hu L, Xiao G et al (2019) Dissecting genetic loci affecting grain morphological traits to improve grain weight via nested association mapping. Theor Appl Genet 132:3115–3128. https://doi.org/10.1007/s00122-019-03410-4
Wang Z, Wu X, Ren Q, Chang X, Li R, Jing R (2010) QTL mapping for developmental behavior of plant height in wheat (Triticum aestivum L.). Euphytica 174:447–458. https://doi.org/10.1007/s10681-010-0166-3
Williams K, Munkvold J, Sorrells M (2013) Comparison of digital image analysis using elliptic Fourier descriptors and major dimensions to phenotype seed shape in hexaploid wheat (Triticum aestivum L.). Euphytica 190:99–116. https://doi.org/10.1007/s10681-012-0783-0
Xin F, Zhu T, Wei S, Han Y, Zhao Y, Zhang D et al (2020) QTL mapping of kernel traits and validation of a major QTL for kernel length-width ratio using SNP and bulked segregant analysis in wheat. Sci Rep 10:1–12. https://doi.org/10.1038/s41598-019-56979-7
Xu D, Wen W, Fu L, Li F, Li J, Xie L et al (2019) Genetic dissection of a major QTL for kernel weight spanning the Rht-B1 locus in bread wheat. Theor Appl Genet 132:3191–3200. https://doi.org/10.1007/s00122-019-03418-w
Xu R, Duan P, Yu H, Zhou Z, Zhang B, Wang R et al (2018) Control of grain size and weight by the OsMKKK10-OsMKK4-OsMAPK6 signaling pathway in rice. Mol Plant 11:860–873. https://doi.org/10.1016/j.molp.2018.04.004
Xu YF, Li SS, Li LH, Ma FF, Fu XY, Shi ZL et al (2017) QTL mapping for yield and photosynthetic related traits under different water regimes in wheat. Mol Breed. https://doi.org/10.1007/s11032-016-0583-7
Yanagawa Y, Sullivan JA, Komatsu S, Gusmaroli G, Suzuki G, Yin J et al (2004) Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev 18:2172–2181. https://doi.org/10.1101/gad.1229504
Yang W, Liu D, Li J, Zhang L, Wei H, Hu X et al (2009) Synthetic hexaploid wheat and its utilization for wheat genetic improvement in China. J Genet Genomics 36:539–546. https://doi.org/10.1016/S1673-8527(08)60145-9
Yang Z, Bai Z, Li X, Wang P, Wu Q, Yang L et al (2012) SNP identification and allelic-specific PCR markers development for TaGW2, a gene linked to wheat kernel weight. Theor Appl Genet 125:1057–1068. https://doi.org/10.1007/s00122-012-1895-6
Zhang D, Wang B, Zhao J, Zhao X, Zhang L, Liu D et al (2014a) ScienceDirect Divergence in homoeolog expression of the grain length-associated gene GASR7 during wheat allohexaploidization. Crop J 3:1–9. https://doi.org/10.1016/j.cj.2014.08.005
Zhang G, Wang Y, Guo Y, Zhao Y, Kong F, Li S (2015) Characterization and mapping of QTLs on chromosome 2D for grain size and yield traits using a mutant line induced by EMS in wheat. Crop J 3:135–144. https://doi.org/10.1016/j.cj.2014.11.002
Zhang L, Zhao YL, Gao LF, Zhao GY, Zhou RH, Zhang BS et al (2012) TaCKX6-D1, the ortholog of rice OsCKX2, is associated with grain weight in hexaploid wheat. New Phytol 195:574–584. https://doi.org/10.1111/j.1469-8137.2012.04194.x
Zhang Y, Liu J, Xia X, He Z (2014b) TaGS-D1, an ortholog of rice OsGS3, is associated with grain weight and grain length in common wheat. Mol Breed 34:1097–1107. https://doi.org/10.1007/s11032-014-0102-7
Zhao JL, Wang HW, Zhang XC, Du XY, Li AF, Kong LR (2015) Association analysis of grain traits with SSR markers between Aegilops tauschii and hexaploid wheat (Triticum aestivum L.). J Integr Agric 14:1936–1948. https://doi.org/10.1016/S2095-3119(15)61070-X
Zhu J (1995) Analysis of conditional genetic efects and variance components in developmental genetics. Genetics 141:1633–1639
Zhu T, Liang C, Meng Z, Li Y, Wu Y, Guo S (2017) PrimerServer : a high-throughput primer design and specificity-checking platform. bioRxiv, pp 1–6
Acknowledgements
We thank the Triticeae Multi-omics Center (http://202.194.139.32/) for providing an integrated platform of tools and genomic data bringing great convenience to our work. Prof. Mujun Yang of Yunnan Academy of Agricultural Sciences is also acknowledged for the help in developing DH population.
Funding
This work is supported by Science and Technology Support Project of Sichuan Province, China (2021YFYZ0027), National Key R&D Program of China (2016YFD0100102), Key Project of Crop Breeding of Sichuan Province (2021YFYZ0002), Science and technology projects of Sichuan Province (2020YFSY0049), and Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA08020205).
Author information
Authors and Affiliations
Contributions
TL undertook the field trials, data analysis and drafted this manuscript. GD assisted in field trials. YS, ZY, YT, JW, JZ, XQ and XP participated in phenotype measurement. WY, JL and ZL developed and provided us the CC population. MY, HZ and JL discussed results and revised the manuscript. HL and WY designed the experiments, guided the entire study, participated in data analysis, discussed results and revised the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
The authors declare that this research has no human and animal participants and that the experiments comply with the current laws of China.
Availability of data and material
All data used in this study were present in the manuscript and supporting materials.
Additional information
Communicated by Xianchun Xia.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, T., Deng, G., Su, Y. et al. Genetic dissection of quantitative trait loci for grain size and weight by high-resolution genetic mapping in bread wheat (Triticum aestivum L.). Theor Appl Genet 135, 257–271 (2022). https://doi.org/10.1007/s00122-021-03964-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-03964-2