Abstract
Background
Grain size is one of key agronomic traits that determine grain yield in rice. Several regulators of grain size have been identified in rice, but the mechanisms that determine grain size and yield remain largely unknown.
Results
Here we characterize a small grain (smg11) mutant in rice, which exhibits small grains, dense panicles and the increased number of grains per panicle. Cloning and sequence analyses of the SMG11 gene reveal that smg11 is a new allele of DWARF2 (D2), which encodes a cytochrome P450 (CYP90D2) involved in brassinosteroid biosynthetic pathway. Overexpression of D2/SMG11 increases grain size and grain weight of wild-type plants. Overexpression of D2/SMG11 at a suitable level also significantly increases grain yield in rice. Cellular analyses indicate that D2/SMG11 controls grain size by promoting cell expansion. Further results reveal that D2/SMG11 influences expression of several known grain size genes involved in the regulation of cell expansion, revealing a novel link between D2/SMG11 and known grain size genes.
Conclusions
SMG11 controls grain size by promoting cell expansion in grain hulls. SMG11 regulates cell expansion, at least in part, by influencing expression of several grain size genes involved in the regulation of cell expansion. The smg11 is a new allele of DWARF2/D2. The suitable expression of SMG11 increases grain size, grain weight and grain yield. Our findings reveal the functions of D2/SMG11 in grain size and grain yield, suggesting that the suitable expression of D2/SMG11 is a promising approach to improve grain yield in rice.
Similar content being viewed by others
Background
Rice is one of the most important cereal crops and the main food source of the global population. Grain yield is coordinately controlled by grain weight, grain number per panicle, panicle number per plant, and grain filling ratio. Grain weight is positively associated with grain size, which is determined by grain length, grain width, and grain thickness. Several factors that regulate grain size have been reported in rice (Zuo and Li, 2014; Li and Li, 2016), but the genetic and molecular mechanisms of grain size control remain largely unknown.
Several factors that regulate grain size by influencing cell proliferation have been described in rice. The major QTL for both grain length and grain weight (GS3) encodes a putative G protein γ subunit and negatively regulates grain size (Fan et al., 2006; Mao et al., 2010). However, its Arabidopsis homolog (AGG3) promotes seed and organ growth by increasing cell proliferation (Li et al., 2012). The major QTL for grain length (qGL3/GL3.1), which encodes a putative protein phosphatase with Kelch-like repeat domain (OsPPKL1), influences grain length by limiting cell proliferation (Hu et al., 2012; Qi et al., 2012; Zhang et al., 2012). The major QTL for grain width and weight (GW2) encodes a RING-type E3 ubiquitin ligase, which restricts cell proliferation in spikelet hulls (Song et al., 2007). Arabidopsis DA2 shares significant similarity with rice GW2 and functions as a negative regulator of seed and organ size in Arabidopsis (Xia et al., 2013). A ubiquitin-related protein encoded by rice SEED WIDTH ON CHROMOSOME 5 (qSW5/GW5) influences grain width by limiting cell proliferation (Shomura et al., 2008; Weng et al., 2008). Higher expression of GW8/OsSPL16 promotes cell division and grain filling, resulting in wide and heavy grains in rice (Wang et al., 2012). GS5, which encodes a putative serine carboxypeptidase, promotes cell proliferation in spikelet hulls (Li et al., 2011). The MAPK pathway has recently reported to control grain size by increasing cell proliferation in spikelet hulls (Duan et al., 2014; Liu et al., 2015). In addition, several genes that regulate grain size by influencing cell expansion have been reported in rice. The major QTL for grain size (GS2) encodes Growth-Regulating Factor 4 (OsGRF4) (Che et al., 2015; Duan et al., 2015; Hu et al., 2015). OsGRF4/GS2 physically interacts with transcriptional coactivators GRF-interacting Factors (GIFs) to regulate grain size by increasing both cell expansion and cell proliferation in spikelet hulls (Duan et al., 2015). The major QTL for grain length and width (GLW7) encodes the transcription factor OsSPL13, which promotes grain growth by increasing cell expansion in spikelet hulls (Si et al., 2016). OsSPL13 associates the promoter of SMALL AND ROUND SEEDS (SRS5) and regulates its expression (Si et al., 2016). The bHLH transcription factors PGL1 and PGL2 promote grain growth by increasing cell expansion in spikelet hulls, while the bHLH transcription factor APG functions antagonistically with PGL1 and PGL2 to influence grain length (Heang and Sassa, 2012a, b). Thus, the transcriptional regulatory factors play important roles in rice grain size control. Several recent studies show that elevated expression of GL7/GW7/SLG7, which encodes a protein homologous to Arabidopsis thaliana LONGIFOLIA1/2, produces slender and long grains (Wang et al., 2015a; Wang et al., 2015b; Zhou et al., 2015). GL7/GW7/SLG7 has been reported to affect grain length and shape by increasing cell elongation in spikelet hulls (Wang et al., 2015b; Zhou et al., 2015), while another study show that GL7/GW7/SLG7 controls grain size by influencing cell proliferation in spikelet hulls (Wang et al., 2015a). Thus, grain size is coordinately determined by cell proliferation and cell expansion in rice.
To further understand the molecular mechanisms that determine grain size, we have previously isolated small grain mutants (smg) in rice (Duan et al., 2014). Here we report that smg11 is a new allele of DWARF2 (D2), which encodes a cytochrome P450 (CYP90D2) involved in brassinosteroid (BR) biosynthetic pathway (Hong et al., 2005). Brassinosteroids play important roles in plant growth and development. Several studies show that genes involved in BR signaling and BR biosynthetic pathways influence seed size in Arabidopsis and rice (Zuo and Li, 2014; Li and Li, 2015, 2016). In this study, our results reveal that SMG11/DWARF2 (D2) positively regulates grain size by promoting cell expansion in spikelet hulls. smg11 mutant produces small grains due to decreased cell expansion, while overexpression of SMG11 causes large grains as a result of increased cell expansion. Further results show that SMG11 affects expression of several known grain size genes involved in the regulation of cell expansion, revealing a novel link between D2/SMG11 and known grain size regulators. A suitable expression of SMG11 also significantly enhances grain yield in rice. Thus, our findings identify the functions of SMG11 in grain size and yield control and give insight into how grain size is determined in rice.
Results
smg11 Produces Small Grains
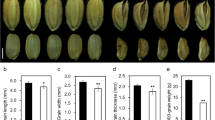
To understand genetic and molecular mechanisms that set the final size of grains, we have previously identified mutants with altered grain size in rice (Duan et al., 2014). The small grain 11 (smg11) mutant was isolated from the ethyl methanesulfonate (EMS)-treated M2 populations of the japonica variety Kuanyejing (KYJ). The smg11 mutant showed obviously smaller grains than KYJ (Fig. 1a, b). The length and width of smg11 grains was significantly decreased compared with that of KYJ grains (Fig. 1c, d). The average length of KYJ and smg11 grains was 6.93 mm and 6.03 mm, respectively. By contrast, the average width of KYJ and smg11 grains was 3.16 mm and 2.95 mm, respectively. In addition, the 1000-grain weight of smg11 was dramatically reduced compared with that of the wild type (Fig. 1e). The 1, 000-grain weight of KYJ was 24.93 g, while the 1, 000-grain weight of smg11 was only 16.86 g. Therefore, these results indicate that the smg11 mutation influences grain size and weight in rice.
The smg11 mutant produces small grains. a Mature paddy rice grains of KYJ and smg11. b Brown rice grains of KYJ and smg11. c The average length of KYJ and smg11 grains. d The average width of KYJ and smg11 grains. e The 1000-grain weight of KYJ and smg11. Values c-e are given as mean ± SD. *P < 0.05; **P < 0.01 compared with the wild type by Student’s t-test. Bars: 2 mm in a and b
smg11 Forms Dense and Erect Panicles With Increased Grain Number
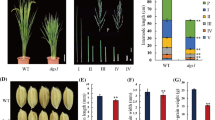
The smg11 plants were obviously shorter than wild-type plants (Fig. 2a, e). The smg11 leaves were more erect than wild-type leaves (Fig. 2a). At the mature stage, the panicles of smg11 exhibited the erect phenotype compared with KYJ panicles (Fig. 2b). The panicles of smg11 were also shorter and denser than those of the wild type (Fig. 2b–c). The panicle axis of smg11 was slightly shorter than that of the wild type (Fig. 2d). These results indicate that the smg11 mutation affects panicle size and shape. We then counted the number of primary and secondary panicle branches. As shown in Fig. 2f, g, the number of primary panicle branches in smg11 was similar to that in KYJ, while the number of secondary panicle branches in smg11 was significantly increased in comparison to that in KYJ. We also observed that the grain number per panicle in smg11 was higher than that in KYJ (Fig. 2c, h). Thus, these results show that the dense panicle phenotype of smg11 was due to a decrease in the length of panicle axis and increases in the secondary panicle branch number and grain number per panicle.
smg11 produces dense and erect panicles. a KYJ (left) and smg11 (right) plants at the mature stage. b c Panicles of KYJ (left) and smg11 (right) at the mature stage. d Distance between panicle neck and each primary branch of KYJ and smg11 panicles. e The average length of KYJ and smg11 panicles. f Total number of KYJ and smg11 primary branches. g Total number of KYJ and smg11 secondary branches. h Grain number per panicle in KYJ and smg11. Values d-h are given as the mean ± SD. *P < 0.05; **P < 0.01 compared with the wild type by Student’s t-test. Bars: 20 cm in a; 5 cm in b-c
smg11 Decreases Cell Expansion in Spikelet Hulls and Influences Expression of Several Known Grain Size Genes
The size of a grain has been known to be restricted by its spikelet hull, which may set an upper limit to final grain size (Li and Li, 2016). The growth of spikelet hulls is coordinately determined by cell proliferation and cell expansion. We therefore examined cells in KYJ and smg11 spikelet hulls. As shown in Fig. 3a–d, outer epidermal cells in smg11 spikelet hulls were significantly shorter and narrower than those in KYJ. Similarly, inner epidermal cells in smg11 spikelet hulls were shorter and narrower than those in KYJ spikelet hulls (Fig. 3e–h). By contrast, the number of epidermal cells in the grain-length direction in smg11 spikelet hulls was similar to that in KYJ spikelet hulls (Additional file 1: Figure S4). These results indicate that the small grain phenotype of smg11 mainly results from the reduced cell expansion in spikelet hulls.
smg11 influences cell expansion. a b SEM analysis of the outer surface of KYJ a and smg11 b lemmas. c The average length of outer epidermal cells in KYJ and smg11 lemmas. d The average width of outer epidermal cells in KYJ and smg11 lemmas. e f SEM analysis of the inner surface of KYJ e and smg11 f lemmas. g The average length of inner epidermal cells in KYJ and smg11 lemmas. h The average width of inner epidermal cells in KYJ and smg11 lemmas. i Expression levels of the indicated genes in KYJ and smg11 panicles. j Expression levels of the indicated genes in KYJ and smg11 panicles. Values c-d; g–j are means ± SD. *P < 0.05; **P < 0.01 compared with the wild type by Student’s t-test. Bars: 100 μm in a, b, e, f
To understand how D2/SMG11 regulates cell expansion in spikelet hulls, we investigated expression of several known grain size genes involved in the regulation of cell expansion, including GS2, GL7, GLW7/SPL13, PGL1, PGL2, APG and SRS5. The transcription factor SPL13/GWL7 has been recently reported to promote expression of SRS5 that encodes alpha-tubulin protein (Si et al., 2016). Overexpression of SPL13 increased grain length by promoting cell elongation in spikelet hulls (Si et al., 2016). Expression levels of both SPL13/GWL7 and SRS5 in smg11 panicles were lower than those in KYJ panicles (Fig. 3i). The bHLH transcription factors PGL1 and PGL2 positively regulate grain length by increasing cell expansion in spikelet hulls (Heang and Sassa, 2012a, b). In contrast, the bHLH transcription factor APG functions antagonistically with PGL1 and PGL2 to influence grain length by influencing cell expansion in spikelet hulls (Heang and Sassa, 2012a, b). As shown in Fig. 3i, expression levels of PGL1 and PGL2 in smg11 were lower than those in KYJ, while expression level of APG in smg11 was higher than that in KYJ. Two studies showed that GL7/GW7/SLG7 regulates grain size by influencing cell expansion in spikelet hulls (Wang et al., 2015b; Zhou et al., 2015), although another study reported that GL7/GW7/SLG7 controls grain size by regulating cell proliferation (Wang et al., 2015a). We observed that expression of GL7/GW7/SLG7 was significantly reduced in smg11 in comparison to that in KYJ (Fig. 3i). The transcription factor GS2 and a putative serine carboxypeptidase GS5 regulate grain size by promoting both cell proliferation and cell expansion (Li et al., 2011; Che et al., 2015; Duan et al., 2015; Hu et al., 2015). Expression levels of GS2 and GS5 in smg11 were similar to those in KYJ (Fig. 3i, j). Furthermore, we detected expression of several grain size genes involved in the regulation of cell proliferation in spikelet hulls, including GW2, GS3 and GL3. As shown in Fig. 3j, expression levels of GW2, GS3 and GL3 in smg11 were not strongly altered in comparison to those in KYJ, further suggesting the role of SMG11 in cell expansion. Taken together, these results suggest that SMG11 regulates grain size, at least in part, by influencing expression of these grain size genes involved in the regulation of cell expansion.
smg11 is a Novel Allele of the DWARF 2 (D2) Gene
The smg11 mutation was identified using the MutMap approach (Abe et al., 2012), which is based on whole-genome resequencing of bulked DNA of F2 segregants. An F2 population of a cross between smg11 and the parental line KYJ were generated. The segregation of F2 progenies showed that the phenotypes of smg11 are determined by a single recessive gene. DNA from 50 F2 individuals that showed the small-grain phenotype was pooled in an equal ratio and subjected to whole-genome resequencing, and DNA from KYJ was resequenced as a control. We obtained a total of 5.6 Gbp of short reads for KYJ and 8.7 Gbp for the bulked F2 plants. 2928 SNPs and 423 INDELs that represent the polymorphisms between the bulked F2 and KYJ were detected. For these SNPs and INDELs, the SNP/INDEL-index (the ratio between the number of reads of a mutant SNP/INDEL and the total number of reads corresponding to the SNP/INDEL) were calculated. The causative SNP/INDEL should be shared by all the mutant F2 plants and therefore has a SNP/INDEL-index = 1. In all, 47 SNPs and 34 INDELs have a SNP/INDEL-index = 1, while only 2 SNPs were identified in exons (Additional file 1: Figure S1). The SNP1 represents a missense mutation of a proline (CCC) codon to a leucine (CTC) in LOC_Os01g10040, while the SNP2 is corresponded to a synonymous mutation. Thus, these results suggest that LOC_Os01g10040 is a good candidate gene for SMG11.
The identity of the SMG11 gene was further confirmed by genetic complementation analysis. A plasmid that contained wild-type gene of LOC_Os01g10040 driven by a Actin promoter (pActin: SMG11) was introduced into the smg11 mutant. We generated 58 transgenic plants and found that transgenic plants showed wild-type phenotypes (Fig. 4d–j). For example, grain length, grain width and 1000-grain weight of pActin: SMG11;smg11 were similar to those of KYJ. Similarly, the secondary branch number and grain number per panicle of pActin: SMG11;smg11 were comparable with those of KYJ. In addition, pActin: SMG11;smg11 transgenic lines exhibited similar plant height and panicle length to KYJ plants (Additional file 1: Figure S2). Therefore, these results indicate that LOC_Os01g10040 is the SMG11 gene.
Identification of the D2/SMG11 gene. a The D2/SMG11 gene structure. The white boxes indicate the exons, and black lines indicate the introns. The start codon (ATG) and the stop codon (TAG) are indicated. The smg11 mutation in the SMG11/D2 gene is shown. b The dCAPS marker is developed according to the smg11 mutation. c The D2/SMG11 protein contains a membrane anchor domain (N1), a proline-rich domain (N2), a dioxygen binding domain (N3), a steroid binding domain (N4) and a heme binding domain (N5). d e Grains of KYJ, smg11, pActin: SMG11;smg11#1 and pActin: SMG11;smg11#2. pActin: SMG11;smg11 is smg11 transformed with pActin: SMG11. f g h Grain length f grain width g and 1000-grain weight h of KYJ, smg11, pActin: SMG11;smg11#1 and pActin: SMG11;smg11#2. i The number of KYJ, smg11, pActin: SMG11;smg11#1 and pActin: SMG11;smg11#2 secondary panicle branches. j Grain number per panicle of KYJ, smg11, pActin: SMG11;smg11#1 and pActin: SMG11;smg11#2. k Quantitative real-time RT-PCR analysis of SMG11/D2 gene expression in developing panicles of 1 cm (YP1) to 20 cm (YP20). Values f–k are given as mean ± SD. *P < 0.05; **P < 0.01 compared with smg11 by Student’s t-test. Bars: 2 mm in d-e
The gene LOC_Os01g10040 encodes a cytochrome P450 protein CYP90D2/DWARF2 (D2), which is involved in the BR biosynthesis pathway (Hong et al., 2005). Expression of SMG11/D2 was detected in developing panicles (Fig. 4k), consistent with the roles of D2/SMG11 in grain size and panicle size control. The smg11 has a C to T transition in codon 74 (CCC/CTC) of LOC_Os01g10040 (Fig. 4a, c), resulting in a Pro/Leu amino acid substitution (Fig. 4c). Sequence alignment of several rice and Arabiodpsis SMG11/D2 homologs showed that Pro in the position 74 is a conserved amino acid (Additional file 1: Figure S3). These results show that smg11 is a novel allele of CYP90D2/D2.
smg11 Affects Expression of Brassinosteroid Biosynthetic and Signaling Genes
It is known that BR signaling mutants or BR-synthetic mutants influence expression levels of BR-synthetic genes as a feedback mechanism (Tong et al., 2009; Duan et al., 2014). We therefore examined expression levels of several BR-synthetic genes in KYJ and smg11 panicles, including DWARF4, DWARF11 and BRD1 (Hong et al., 2002; Mori et al., 2002; Tanabe et al., 2005; Sakamoto et al., 2006). As shown in Fig. 5, expression levels of DWARF4, DWARF11 and BRD1 in smg11 panicles were higher than those in KYJ panicles. It is possible that the mutation in SMG11/D2 resulted in the feedback up-regulation of BR biosynthetic genes in rice.
We then asked whether SMG11 influences expression of BR-signaling genes involved in the regulation grain size, such as OsBRI1, GSK2, OsBZR1, and BU1 (Yamamuro et al., 2000; Tanaka et al., 2009; Tong et al., 2012; Zhu et al., 2015). We performed quantitative real-time RT-PCR analysis to investigate their expression levels in KYJ and smg11 panicles. As shown in Fig. 5, expression levels of BRI1 and BU1 in smg11 panicles were lower than those in KYJ panicles, while expression levels of GSK2 and OsBZR1 in smg11 panicles were not significantly different from those in KYJ panicles. It is plausible that the mutation in SMG11/D2 might decrease BR signaling or responses by repressing expression of BR signaling genes (e.g. OsBRI1 and BU1), resulting in small grains.
Overexpression of SMG11 Increases Grain Size due to Large Cells in Spikelet Hulls
To further understand roles of SMG11 in grain size control and its potential application in rice yield improvement, we expressed the SMG11 gene driven by a Actin promoter (pActin: SMG11) in a japonica variety Zhonghua 11 (ZH11) and generated 29 transgenic plants. Transgenic plants produced longer and wider grains than ZH11 (Fig. 6a, b, g, h). The 1000-grain weight in pActin: SMG11 transgenic plants was also significantly increased compared with that in ZH11 (Fig. 6i). We further examined expression levels of SMG11/D2 in these transgenic plants (Fig. 6j). Expression levels of SMG11/D2 were positively correlated with the grain size and weight phenotypes of transgenic plants.
Overexpression of D2/SMG11 increases grain size due to large cells in spikelet hulls. a b Grains of ZH11, pActin: SMG11#1, pActin: SMG11#2 and pActin: SMG11#3. pActin: SMG11 is ZH11 transformed with pActin: SMG11. c d SEM analysis of the outer surface of ZH11 c and pActin: SMG11#2 d lemmas. e, f SEM analysis of the inner surface of ZH11 e and pActin: SMG11#2 f lemmas. g h i Grain length g grain width h and 1000-grain weight i of ZH11, pActin:SMG11#1, pActin:SMG11#2 and pActin:SMG11#3. j Expression levels of SMG11/D2 in ZH11, pActin:SMG11#1, pActin:SMG11#2 and pActin:SMG11#3 panicles. k The average length of outer epidermal cells in ZH11 and pActin:SMG11#2 lemmas. l The average width of outer epidermal cells in ZH11 and pActin:SMG11#2 lemmas. m The average length of inner epidermal cells in ZH11 and pActin:SMG11#2 lemmas. n The average width of inner epidermal cells in ZH11 and pActin:SMG11#2 lemmas. Values g–n are given as mean ± SD. *P < 0.05 **P < 0.01 compared with ZH11 by Student’s t-test. Bars: 2 mm in a-b 100 μm in c–f
To investigate cellular basis for the large grain phenotype of pActin: SMG11 transgenic plants, we examined cell size of ZH11 and pActin: SMG11 spikelet hulls. As shown in Fig. 6c, d, k, l, outer epidermal cells in pActin: SMG11 spikelet hulls were significantly longer and wider than those in ZH11. Similarly, inner epidermal cells in pActin: SMG11 spikelet hulls were longer and wider than those in ZH11 spikelet hulls (Fig. 6e, f, m, n). These results indicate that SMG11 promotes grain growth by increasing cell expansion in spikelet hulls.
A Suitable Expression of SMG11 Increases Grain Yield in Rice
As pActin: SMG11 transgenic plants produced large and heavy grains, we asked whether overexpression of SMG11 could increase grain yield in rice. We therefore investigated grain yield per plant of pActin: SMG11 transgenic plants with different expression levels of SMG11. As shown in Fig. 6j, relative expression levels of SMG11 in pActin: SMG11#1, pActin: SMG11#2 and pActin: SMG11#3 transgenic plants were 31.25, 177.42 and 246.66 folds higher than those in ZH11, respectively. pActin: SMG11#1 transgenic plants exhibited higher yield per plant than ZH11 plants (Fig. 7a). In contrast, pActin: SMG11#2 and #3 transgenic plants decreased grain yield per plant (Fig. 7a), although they produced large and heavy grains (Fig. 6g–i). These results suggest that the effect of D2/SMG11 on grain yield depends on its expression levels. To address why different pActin: SMG11 lines showed opposite effects on grain yield, we investigated panicle branch number and grain number per panicle of pActin: SMG11 transgenic lines. As shown in Fig. 7b–d, the primary panicle branch number, the secondary panicle branch number and grain number per panicle in pActin: SMG11#1 were similar to those in ZH11, while the secondary panicle branch number and grain number per panicle in pActin: SMG11#2 and pActin: SMG11#3 were significantly reduced in comparison to those in ZH11. Thus, pActin: SMG11#1 plants had normal panicle branch number as well as large and heavy grains, resulting in high grain yield. These results suggest that optimizing SMG11 expression could be utilized to increase grain yield in rice.
The suitable expression of SMG11 increases grain yield. a Grain yield per plant of ZH11, pActin:SMG11#1, pActin:SMG11#2 and pActin:SMG11#3. b–d The primary panicle branch number b the secondary panicle branch number c and grain number per panicle d of ZH11, pActin:SMG11#1, pActin:SMG11#2 and pActin:SMG11#3. Values a–d are given as mean ± SD. *P < 0.05 **P < 0.01 compared with ZH11 by Student’s t test
Discussion
Grain size is one of important agronomic traits in crops. Grain size is determined by grain length, grain width and grain thickness. Several factors that control grain size have been identified in rice, but the mechanisms that control grain size remain largely unknown. It is also a great challenge to improve rice yield using these grain size regulators. In this study, we report that smg11 is a novel allele of D2 (Hong et al., 2003). Our results show that SMG11 promotes grain growth by increasing cell expansion in spikelet hulls. SMG11 controls grain size, at least in part, by influencing expression of several known grain size genes involved in the regulation of cell expansion. Further results reveal that a suitable expression of SMG11 increases grain size, grain weight and grain yield, suggesting that it is a promising target for rice yield improvement.
The smg11 mutant produced small grains and dense, erect and short panicles, indicating that SMG11 influences grain and panicle size. The secondary panicle branches and grain number per panicle in smg11 were increased compared with KYJ, suggesting a possible balance mechanism between grain size and grain number. The smg11 mutation occurred in the cytochrome P450 CYP90D2/D2, which is involved in BR biosynthesis (Hong et al., 2005). smg11 is a novel allele of D2/CYP90D2. The smg11 also showed similar phenotypes to BR defective mutants, such as reduced leaf angle and small grains (Additional file 1: Figures S5 and S6a). Exogenous application of BL rescued the leaf angle phenotype of smg11 (Additional file 1: Fig. S6), further suggesting that the smg11 mutation influences BR biosynthesis. Although several rice d2 alleles have been previously reported (Hong et al., 2003; Hong et al., 2005; Li et al., 2013), none of these alleles was reported to increase the panicle branches and grain number per panicle. Consistent with the role of SMG11 in panicle branch and grain number control, expression of SMG11 complemented the panicle branch and grain number phenotypes of smg11, and strong expression of SMG11 decreased panicle branches and grain number per panicle.
The d2 alleles have been shown to produce small grains (Hong et al., 2003; Hong et al., 2005; Li et al., 2013), but how D2 influences grain size is almost unknown. Our results showed that smg11 had small grains due to short and narrow cells in spikelet hulls, indicating that SMG11 regulates grain size by promoting cell expansion in both grain-length and grain-width directions. Several factors that control grain size by regulating cell expansion have been reported in rice. For example, the transcription factor SPL13/GWL7 binds to the promoter regions of SRS5 and promotes its expression (Si et al., 2016). Overexpression of SPL13 increased grain length by promoting cell elongation in spikelet hulls. Interestingly, we found that expression levels of both SPL13/GWL7 and SRS5 in smg11 panicles were decreased compared with those in KYJ panicles. Elevated expression of GL7, which might be caused by increased copy number or mutations in the promoter, caused long grains due to increased cell elongation in spikelet hulls (Wang et al., 2015b; Zhou et al., 2015), although another study showed that GL7 promotes cell proliferation in the grain-length direction (Wang et al., 2015a). Interestingly, the smg11 mutation also reduced the expression of GL7. The bHLH transcription factors PGL1 and PGL2 have been known to positively regulate grain length by increasing cell expansion in spikelet hulls (Heang and Sassa, 2012a, b). In contrast, the bHLH transcription factor APG functions antagonistically with PGL1 to influence grain length (Heang and Sassa, 2012a, b). Consistent with the roles of PGL1, PGL2 and APG, the smg11 mutation decreased expression of PGL1 and PGL2 and increased expression of APG. D2/SMG11 has been known to be involved in BR biosynthesis (Hong et al., 2005). Several BR mutants have been reported to form small grains in rice, suggesting that BRs play key roles in grain size control. It is possible that BRs might regulate grain size by influencing expression of these grain size genes involved in the regulation of cell expansion, such as SPL13, GL7, PGL1, PGL2 and APG. Consistent with this notion, PGL1 and APG have been proposed to influence BR signaling pathway (Heang and Sassa, 2012a, b). It will be interesting to test whether other BR-deficient mutants might affect expression of these grain size genes in the future.
BRs have been proposed to have potential applications in improving crop yields. In rice, a brassinosteroid-deficient mutant osdwarf4-1 was reported to be associated with the increased grain yield under conditions of dense planting, even without extra fertilizer (Sakamoto et al., 2006). Expression of OsDWARF4 driven by a specific promoter also increased grain yield in rice (Wu et al., 2008). In contrast, overexpression of OsDWARF4 driven by a constitutive promoter caused defects in plant growth, resulting in a decrease in grain production. Therefore, it is possible that an optimized expression of BR-related genes could increase grain yield in rice. In this study, we found that overexpression of SMG11 increased grain size and weight of wild-type plants, supporting that SMG11 is a positive factor of gain size and weight and also suggesting that SMG11 might be a good target for rice yield improvement. As we expected, pActin: SMG11#1 transgenic plants with a suitable level of SMG11 expression increased grain yield per plant. However, pActin: SMG11#2 and pActin: SMG11#3 transgenic plants with much strong expression of SMG11 reduced grain yield due to decreases in panicle branches and grain number per panicle. Therefore, the fine tuning of SMG11 expression will be a promising strategy for increasing grain size and improving grain yield in rice.
Conclusions
The rice smg11 mutant shows small grains, dense panicles and the increased grain number. The smg11 is a new allele of DWARF2 (D2), which encodes a cytochrome P450 (CYP90D2). The SMG11 controls grain size by promoting cell expansion in grain hulls. SMG11 regulates cell expansion, at least in part, by influencing expression of several grain size genes involved in the regulation of cell expansion, revealing a novel link between D2/SMG11 and known grain size genes. The suitable expression of SMG11 increases grain size, grain weight and grain yield in rice. Our findings define the functions of D2/SMG11 in grain size and grain yield, suggesting that an optimized expression of D2/SMG11 is a promising approach to improve grain yield in rice.
Methods
Plant Materials and Growth Conditions
The smg11 mutant was isolated from the japonica variety Kuanyejing (KYJ) mutagenized with EMS. Rice plants were grown in the experimental fields of Hainan (Lingshui, Hainan), the China National Rice Research Institute (Hangzhou, China) and Institute of Genetics and Developmental Biology (Beijing, China) under natural growth conditions.
Morphological and cellular analysis
Mature grains were observed under a Leica microscope (LEICA S8APO; Leica microsystems, Wetzlar, Germany) and photographed using a Leica CCD (DFC420). Grain length and width were measured using Image J software. Grain weight was determined by weighting 1000 dry grains using an electronic analytical balance (Mettler Toledo AL104, China). The weights of three replicates were measured for each grain lot.
The size of epidermal cells in spikelet hulls were investigated using a scanning electron microscope. The samples were fixed in FAA solution (glacial acetic acid: formalin: 50% ethanol; 1:1:18) at 4 °C overnight, dehydrated in a graded ethanol series, and substituted with 100% ethanol. The critical-point drier (HITACHI HCP-2) was used to dry samples. The samples were dissected under a microscope (LEICA S8APO; Leica microsystems, Wetzlar, Germany), sputter-coated with platinum and observed using a scanning electron microscope (HITACHI S-3000 N; Hitachi High-Technologies Corporation, Tokyo, Japan). Cell size was measured using Image J software.
Molecular cloning of D2/SMG11
To identify smg11 mutation, we generated an F2 population from a cross between smg11 and KYJ. We selected plants with smg11 phenotypes from this F2 population and pooled their genomic DNA for the whole genome sequencing. We obtained a total of 5.6 Gbp of short reads for KYJ and 8.7 Gbp for the bulked F2. Then these short reads were alignment to the reference genome sequence (Nipponbare), and 2928 SNPs and 423 INDELs which are specific for the bulked F2 were identified. In these SNP/INDELs position, sequence of KYJ is same to Nipponbare. Thus, these SNP/INDELs represent the polymorphism between the bulked F2 and KYJ. For these SNPs and INDELs, the SNP/INDEL-index (the ratio between the number of reads of a mutant SNP/INDEL and the total number of reads corresponding to the SNP/INDEL) were calculated. The causative SNP/INDEL should be shared by all the mutant F2 plants and therefore has a SNP/INDEL-index = 1. Among them, 47 SNPs and 34 INDELs have a SNP/INDEL-index = 1, while only 2 SNPs were identified in extron. The SNP1 represents a missense mutation of a proline (CCC) codon to a leucine (CTC) in LOC_Os01g10040, while the SNP2 is corresponded to a synonymous mutation. Thus, these results suggested that LOC_Os01g10040 might represent the SMG11 gene.
Plasmid construction and plant transformation
The pActin: SMG11 construct was conducted using a PCR-based Gateway system. The SMG11/D2 gene was amplified using the primers SMG11-F and SMG11-R (Additional file 2: Table S1). PCR products were subcloned into the pCR8/GW/TOPO TA cloning vector (Invitrogen, Carlsbad, CA, USA). The SMG11 gene was further cloned into the pIPKb003 vector with the Actin promoter to generate the plasmid pActin: SMG11. The pActin: SMG11 plasmid was introduced into Agrobacterium strain EHA105, and Zhonghua 11 (ZH11) and smg11 were transformed according to a previous method (Hiei et al., 1994).
RNA extraction and quantitative real-time RT-PCR
Total RNA was isolated from young panicles of KYJ and smg11 using plant RNeasy Mini Kit according to the manufacture manual (TIANGEN, Beijing, China). RNAs were quantified and reversely transcripted into cDNA using SuperScript III Reverse Transcriptase (Invitrogen). First-strand cDNA was synthesized from 3 μg of total RNAs. Reverse transcription was performed at 50 °C for 1 h and 70 °C for 15 min. Quantitative real-time RT-PCR analysis was performed using the CFX96 Real-Time PCR detection system (Bio-Rad) and SYBR Green I Master (Roche, Mannheim, Germany). ACTIN1 was used as an internal control. The primers used in quantitative real-time RT-PCR were described in Additional file 2: Table S1.
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012) Genome Sequencing Reveals Agronomically Important Loci in Rice Using MutMap. Nat Biotechnol 30:174–178
Che R, Tong H, Shi B, Liu Y, Fang S, Liu D, Xiao Y, Hu B, Liu L, Wang H, Zhao M, Chu C (2015) Control of Grain Size and Rice Yield by GL2-Mediated Brassinosteroid Responses. Nat Plants 2:15195
Duan P, Rao Y, Zeng D, Yang Y, Xu R, Zhang B, Dong G, Qian Q, Li Y (2014) SMALL GRAIN 1, Which Encodes a Mitogen-Activated Protein Kinase Kinase 4, Influences Grain Size in Rice. Plant J 77:547–557
Duan P, Ni S, Wang J, Zhang B, Xu R, Wang Y, Chen H, Zhu X, Li Y (2015) Regulation of OsGRF4 by OsmiR396 Controls Grain Size and Yield in Rice. Nat Plants 2:15203
Fan C, Xing Y, Mao H, Lu T, Han B, Xu C, Li X, Zhang Q (2006) GS3, a Major QTL for Grain Length and Weight and Minor QTL for Grain Width and Thickness in Rice, Encodes a Putative Transmembrane Protein. Theor Appl Genet 112:1164–1171
Heang D, Sassa H (2012a) An Atypical bHLH Protein Encoded by POSITIVE REGULATOR OF GRAIN LENGTH 2 is Involved in Controlling Grain Length and Weight of Rice Through Interaction With a Typical bHLH Protein APG. Breed Sci 62:133–141
Heang D, Sassa H (2012b) Antagonistic Actions of HLH/bHLH Proteins are Involved in Grain Length and Weight in Rice. Plos One 7:e31325
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient Transformation of Rice (Oryza Sativa L.) Mediated by Agrobacterium and Sequence Analysis of the Boundaries of the T-DNA. Plant J 6:271–282
Hong Z, Ueguchi-Tanaka M, Fujioka S, Takatsuto S, Yoshida S, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M (2005) The Rice Brassinosteroid-Deficient dwarf2 Mutant, Defective in the Rice Homolog of Arabidopsis DIMINUTO/DWARF1, is Rescued by the Endogenously Accumulated Alternative Bioactive Brassinosteroid, Dolichosterone. Plant Cell 17:2243–2254
Hong Z, Ueguchi-Tanaka M, Umemura K, Uozu S, Fujioka S, Takatsuto S, Yoshida S, Ashikari M, Kitano H, Matsuoka M (2003) A Rice Brassinosteroid-Deficient Mutant, Ebisu Dwarf (d2), is Caused by a Loss of Function of a new Member of Cytochrome P450. Plant Cell 15:2900–2910
Hong Z, Ueguchi-Tanaka M, Shimizu-Sato S, Inukai Y, Fujioka S, Shimada Y, Takatsuto S, Agetsuma M, Yoshida S, Watanabe Y, Uozu S, Kitano H, Ashikari M, Matsuoka M (2002) Loss-of-Function of a Rice Brassinosteroid Biosynthetic Enzyme, C-6 Oxidase, Prevents the Organized Arrangement and Polar Elongation of Cells in the Leaves and Stem. Plant J 32:495–508
Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D, Kang S, Zhu L, Dong G, Guo L, Zeng D, Zhang G, Xie L, Xiong G, Li J, Qian Q (2015) A Rare Allele of GS2 Enhances Grain Size and Grain Yield in Rice. Mol Plant 8:1455–1465
Hu Z, He H, Zhang S, Sun F, Xin X, Wang W, Qian X, Yang J, Luo X (2012) A Kelch Motif-Containing Serine/Threonine Protein Phosphatase Determines the Large Grain QTL Trait in Rice. J Integr Plant Biol 54:979–990
Li H, Jiang L, Youn JH, Sun W, Cheng Z, Jin T, Ma X, Guo X, Wang J, Zhang X, Wu F, Wu C, Kim SK, Wan J (2013) A Comprehensive Genetic Study Reveals a Crucial Role of CYP90D2/D2 in Regulating Plant Architecture in Rice (Oryza Sativa). New Phytol 200:1076–1088
Li N, Li Y (2015) Maternal Control of Seed Size in Plants. J Exp Bot 66:1087–1097
Li N, Li Y (2016) Signaling Pathways of Seed Size Control in Plants. Curr Opin Plant Biol 33:23–32
Li S, Liu Y, Zheng L, Chen L, Li N, Corke F, Lu Y, Fu X, Zhu Z, Bevan MW, Li Y (2012) The Plant-Specific G Protein Gamma Subunit AGG3 Influences Organ Size and Shape in Arabidopsis Thaliana. New Phytol 194:690–703
Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J, He Y, Zhang Q (2011) Natural Variation in GS5 Plays an Important Role in Regulating Grain Size and Yield in Rice. Nat Genet 43:1266–1269
Liu S, Hua L, Dong S, Chen H, Zhu X, Jiang J, Zhang F, Li Y, Fang X, Chen F (2015) OsMAPK6, a Mitogen-Activated Protein Kinase, Influences Rice Grain Size and Biomass Production. Plant J 84:672–681
Mao H, Sun S, Yao J, Wang C, Yu S, Xu C, Li X, Zhang Q (2010) Linking Differential Domain Functions of the GS3 Protein to Natural Variation of Grain Size in Rice. Proc Natl Acad Sci U S A 107:19579–19584
Mori M, Nomura T, Ooka H, Ishizaka M, Yokota T, Sugimoto K, Okabe K, Kajiwara H, Satoh K, Yamamoto K, Hirochika H, Kikuchi S (2002) Isolation and Characterization of a Rice Dwarf Mutant With a Defect in Brassinosteroid Biosynthesis. Plant Physiol 130:1152–1161
Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang L, Gao JP, Lin HX (2012) The Novel Quantitative Trait Locus GL3.1 Controls Rice Grain Size and Yield by Regulating Cyclin-T1;3. Cell Res 22:1666–1680
Sakamoto T, Morinaka Y, Ohnishi T, Sunohara H, Fujioka S, Ueguchi-Tanaka M, Mizutani M, Sakata K, Takatsuto S, Yoshida S, Tanaka H, Kitano H, Matsuoka M (2006) Erect Leaves Caused by Brassinosteroid Deficiency Increase Biomass Production and Grain Yield in Rice. Nat Biotechnol 24:105–109
Shomura A, Izawa T, Ebana K, Ebitani T, Kanegae H, Konishi S, Yano M (2008) Deletion in a Gene Associated With Grain Size Increased Yields During Rice Domestication. Nat Genet 40:1023–1028
Si L, Chen J, Huang X, Gong H, Luo J, Hou Q, Zhou T, Lu T, Zhu J, Shangguan Y, Chen E, Gong C, Zhao Q, Jing Y, Zhao Y, Li Y, Cui L, Fan D, Lu Y, Weng Q, Wang Y, Zhan Q, Liu K, Wei X, An K, An G, Han B (2016) OsSPL13 Controls Grain Size in Cultivated Rice. Nat Genet 48:447–456
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX (2007) A QTL for Rice Grain Width and Weight Encodes a Previously Unknown RING-Type E3 Ubiquitin Ligase. Nat Genet 39:623–630
Tanabe S, Ashikari M, Fujioka S, Takatsuto S, Yoshida S, Yano M, Yoshimura A, Kitano H, Matsuoka M, Fujisawa Y, Kato H, Iwasaki Y (2005) A Novel Cytochrome P450 is Implicated in Brassinosteroid Biosynthesis via the Characterization of a Rice Dwarf Mutant, dwarf11, With Reduced Seed Length. Plant Cell 17:776–790
Tanaka A, Nakagawa H, Tomita C, Shimatani Z, Ohtake M, Nomura T, Jiang CJ, Dubouzet JG, Kikuchi S, Sekimoto H, Yokota T, Asami T, Kamakura T, Mori M (2009) BRASSINOSTEROID UPREGULATED1, Encoding a Helix-Loop-Helix Protein, is a Novel Gene Involved in Brassinosteroid Signaling and Controls Bending of the Lamina Joint in Rice. Plant Physiol 151:669–680
Tong H, Liu L, Jin Y, Du L, Yin Y, Qian Q, Zhu L, Chu C (2012) DWARF AND LOW-TILLERING Acts as a Direct Downstream Target of a GSK3/SHAGGY-Like Kinase to Mediate Brassinosteroid Responses in Rice. Plant Cell 24:2562–2577
Tong H, Jin Y, Liu W, Li F, Fang J, Yin Y, Qian Q, Zhu L, Chu C (2009) DWARF AND LOW-TILLERING, a new Member of the GRAS Family, Plays Positive Roles in Brassinosteroid Signaling in Rice. Plant J 58:803–816
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X (2012) Control of Grain Size, Shape and Quality by OsSPL16 in Rice. Nat Genet 44:950–954
Wang S, Li S, Liu Q, Wu K, Zhang J, Wang Y, Chen X, Zhang Y, Gao C, Wang F, Huang H, Fu X (2015a) The OsSPL16-GW7 Regulatory Module Determines Grain Shape and Simultaneously Improves Rice Yield and Grain Quality. Nat Genet 47:949–954
Wang Y, Xiong G, Hu J, Jiang L, Yu H, Xu J, Fang Y, Zeng L, Xu E, Ye W, Meng X, Liu R, Chen H, Jing Y, Zhu X, Li J, Qian Q (2015b) Copy Number Variation at the GL7 Locus Contributes to Grain Size Diversity in Rice. Nat Genet 47:944–948
Weng J, Gu S, Wan X, Gao H, Guo T, Su N, Lei C, Zhang X, Cheng Z, Guo X, Wang J, Jiang L, Zhai H, Wan J (2008) Isolation and Initial Characterization of GW5, a Major QTL Associated With Rice Grain Width and Weight. Cell Res 18:1199–1209
Wu CY, Trieu A, Radhakrishnan P, Kwok SF, Harris S, Zhang K, Wang J, Wan J, Zhai H, Takatsuto S, Matsumoto S, Fujioka S, Feldmann KA, Pennell RI (2008) Brassinosteroids Regulate Grain Filling in Rice. Plant Cell 20:2130–2145
Xia T, Li N, Dumenil J, Li J, Kamenski A, Bevan MW, Gao F, Li Y (2013) The Ubiquitin Receptor DA1 Interacts With the E3 Ubiquitin Ligase DA2 to Regulate Seed and Organ Size in Arabidopsis. Plant Cell 25:3347–3359
Yamamuro C, Ihara Y, Wu X, Noguchi T, Fujioka S, Takatsuto S, Ashikari M, Kitano H, Matsuoka M (2000) Loss of Function of a Rice Brassinosteroid insensitive1 Homolog Prevents Internode Elongation and Bending of the Lamina Joint. Plant Cell 12:1591–1606
Zhang X, Wang J, Huang J, Lan H, Wang C, Yin C, Wu Y, Tang H, Qian Q, Li J, Zhang H (2012) Rare Allele of OsPPKL1 Associated With Grain Length Causes Extra-Large Grain and a Significant Yield Increase in Rice. Proc Natl Acad Sci U S A 109:21534–21539
Zhou Y, Miao J, Gu H, Peng X, Leburu M, Yuan F, Gao Y, Tao Y, Zhu J, Gong Z, Yi C, Gu M, Yang Z, Liang G (2015) Natural Variations in SLG7 Regulate Grain Shape in Rice. Genetics 201:1591–1599
Zhu X, Liang W, Cui X, Chen M, Yin C, Luo Z, Zhu J, Lucas WJ, Wang Z, Zhang D (2015) Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein. Plant J 82:570–581
Zuo J, Li J (2014) Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu Rev Genet 48:99–118
Acknowledgements
We thank members of the Yunhai laboratory for helpful discussions.
Funding
This work was supported by grants from the National Basic Research Program of China (2013CBA01401; 2016YFD0100500), the National Natural Science Foundation of China (91535203; 31160278), the Ministry of Agriculture of China (2016ZX08009-003), Key Science and Technology Project of Hainan Province (ZDZX2013023) and the Platform Construction Programs of Key Laboratory and Engineering Technology Research Center. The funding agencies have no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Authors’ Contributions
NF, RX, YLUO and YLI designed experiments. NF performed plasmid construction and phenotypic analyses of smg11 and transgenic plants, Quantitative real-time RT-PCR and cell size analysis. RX isolated smg11 mutant and identified the candidate gene for SMG11. BZ performed rice transformation. PD and LH helped to do field experiment. NF, RX, YLUO and YLI analyzed data. NF and YLI prepared Figures. NF, RX and YLI wrote article. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Additional Files
Additional file 1: Figure S1.
Identification of the smg11 mutation. Figure S2. Overexpression of SMG11 complements the phenotypes of smg11. Figure S3. Alignment of D2/SMG11 homologs in rice and Arabidopsis. Figure S4. Effect of SMG11 on cell number in spikelet hulls. Figure S5. smg11 influences leaf angle. Figure S6. Exogenous application of BL rescues the leaf angle phenotype of smg11. (PDF 352 kb)
Additional file 2: Table S1.
Primers used in this study (DOCX 19 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Fang, N., Xu, R., Huang, L. et al. SMALL GRAIN 11 Controls Grain Size, Grain Number and Grain Yield in Rice. Rice 9, 64 (2016). https://doi.org/10.1186/s12284-016-0136-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12284-016-0136-z