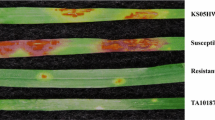
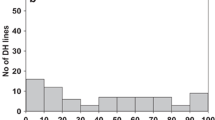
Abstract
Key message
Markers linked to stem rust resistance gene Sr47 were physically mapped in three small Aegilops speltoides chromosomal bins. Five markers, including two PCR-based SNP markers, were validated for marker-assisted selection.
Abstract
In durum wheat (Triticum turgidum subsp. durum), the gene Sr47 derived from Aegilops speltoides conditions resistance to race TTKSK (Ug99) of the stem rust pathogen (Puccinia graminis f. sp. tritici). Sr47 is carried on small interstitial translocation chromosomes (Ti2BL-2SL-2BL·2BS) in which the Ae. speltoides chromosome 2S segments are divided into four bins in genetic stocks RWG35, RWG36, and RWG37. Our objective was to physically map molecular markers to bins and to determine if any of the molecular markers would be useful in marker-assisted selection (MAS). Durum cultivar Joppa was used as the recurrent parent to produce three BC2F2 populations. Each BC2F2 plant was genotyped with markers to detect the segment carrying Sr47, and stem rust testing of BC2F3 progeny with race TTKSK confirmed the genotyping. Forty-nine markers from published sources, four new SSR markers, and five new STARP (semi-thermal asymmetric reverse PCR) markers, were evaluated in BC2F2 populations for assignment of markers to bins. Sr47 was mapped to bin 3 along with 13 markers. No markers were assigned to bin 1; however, 7 and 13 markers were assigned to bins 2 and 4, respectively. Markers Xrwgs38a, Xmag1729, Xwmc41, Xtnac3119, Xrwgsnp1, and Xrwgsnp4 were found to be useful for MAS of Sr47. However, STARP markers Xrwgsnp1 and Xrwgsnp4 can be used in gel-free systems, and are the preferred markers for high-throughput MAS. The physical mapping data from this study will also be useful for pyramiding Sr47 with other Sr genes on chromosome 2B.







Similar content being viewed by others
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Bariana HS, Hayden MJ, Ahmed NU, Bell JA, Sharp PJ, McIntosh RA (2001) Mapping of durable adult plant and seedling resistances to stripe rust and stem rust diseases in wheat. Aust J Agric Res 52:1247–1255
Chao S, Elias E, Benscher D, Ishikawa G, Huang Y-F, Saito M, Nakamura T, Xu S, Faris J, Sorrells M (2016) Genetic mapping of major-effect seed dormancy quantitative trait loci on chromosome 2B using recombinant substitution lines in tetraploid wheat. Crop Sci 56:59–72
Crossa J, Burgueño J, Dreisigacker S, Vargas M, Herrera-Foessel SA, Lillemo M, Singh RP, Trethowan R, Warburton M, Franco J, Reynolds M, Crouch JH, Ortiz R (2007) Association analysis of historical bread wheat germplasm using additive covariance of relatives and population structure. Genetics 177:1889–1913
Dobrovolskaya O, Boeuf C, Salse J, Pont C, Sourdille P, Bernard M, Salina E (2011) Microsatellite mapping of Ae. speltoides and map-based comparative analysis of the S, G, and B genomes of Triticeae species. Theor Appl Genet 123:1145–1157
Elias EM, Manthey FA (2016) Registration of ‘Joppa’ durum wheat. J Plant Reg 10:139–144
Elias EM, Manthey FA, AbuHammad WA (2015) Registration of ‘Carpio’ durum wheat. J Plant Reg 9:78–82
Faris JD, Xu SS, Cai X, Friesen TL, Jin Y (2008) Molecular and cytogenetic characterization of a durum wheat–Aegilops speltoides chromosome translocation conferring resistance to stem rust. Chromosome Res 16:1097–1105
Fernández-Calvín B, Orellana J (1994) Metaphase I-bound arms frequency and genome analysis in wheat–Aegilops hybrids. 3. Similar relationships between the B genome of wheat and S or Sl genomes of Ae. speltoides, Ae. longissima and Ae. sharonenesis. Theor Appl Genet 88:1043–1049
Hossain KG, Riera-Lizarazu O, Kalavacharla V, Vales MI, Maan SS, Kianian SF (2004) Radiation hybrid mapping of the species cytoplasm-specific (scs ae) gene in wheat. Genetics 168:415–423
Jin Y, Singh RP (2006) Resistance in U.S. wheat to recent eastern African isolates of Puccinia graminis f. sp. tritici with virulence to resistance gene Sr31. Plant Dis 90:476–480
Jin Y, Singh RP, Ward R, Wanyera RW, Kinyua R, Njau P, Fetch T, Pretorius ZA, Yahyaoui A (2007) Characterization of seedling infection types and adult plant infection responses of monogenic Sr gene lines to race TTKS of Puccinai graminis f. sp. tritici. Plant Dis 91:1096–1099
Jin Y, Szabo LJ, Pretorius ZA, Singh RP, Ward R, Fetch T Jr (2008) Detection of virulence to resistance gene Sr24 within race TTKS of Puccinai graminis f. sp. tritici. Plant Dis 92:923–926
Jin Y, Szabo LJ, Rouse MN, Fetch T Jr, Pretorius ZA, Wanyera R, Njau P (2009) Detection of virulence to resistance gene Sr36 within the TTKS race lineage of Puccinai graminis f. sp. tritici. Plant Dis 93:367–370
Kalavacharla V, Hossain K, Gu Y, Riera-Lizarazu O, Vales MI, Bhamidimarri S, Gonzalez-Hernandez JL, Maan SS, Kianian SF (2006) High-resolution radiation hybrid map of wheat chromosome 1D. Genetics 173:1089–1099
Kielsmeier-Cook J, Danilova TV, Friebe B, Rouse MN (2015) Resistance to the Ug99 race group of Puccinia graminis f. sp. tritici in wheat-intra/intergeneric hybrid derivatives. Plant Dis 99:1317–1325
Kilian B, Özkan H, Deusch O, Effgen S, Brandolini A, Kohl J, Martin W, Salamini F (2007) Independent wheat B and G genome origins in outcrossing Aegilops progenitor haplotypes. Mol Biol Evol 24:217–227
Klindworth DL, Xu SS (2008) Development of a set of stem rust susceptible D-genome disomic substitutions based on Rusty durum. In: Appels R, Eastwood R, Lagudah E, Langridge P, MacKay M, McIntyre L, Sharp P (eds) Proceedings of the 11th International Wheat Genetics Symposium, vol 2, Brisbane, Queensland, Australia. 24–29 August, 2008. Sydney University Press, Sydney, pp 367–369
Klindworth DL, Miller JD, Xu SS (2006) Registration of Rusty durum wheat. Crop Sci 46:1012–1013
Klindworth DL, Miller JD, Jin Y, Xu SS (2007) Chromosomal locations of genes for stem rust resistance in monogenic lines derived from tetraploid wheat accession ST464. Crop Sci 47:1441–1450
Klindworth DL, Niu Z, Chao S, Friesen TL, Jin Y, Faris JD, Cai X, Xu SS (2012) Introgression and characterization of a goatgrass gene for a high level of resistance to Ug99 stem rust in tetraploid wheat. Genes Genomes Genetics 2:665–673
Kumar S, Goyal A, Mohan A, Balyan HS, Gupta PK (2013) An integrated physical map of simple sequence repeats in bread wheat. Aust J Crop Sci 7:460–468
Letta T, Olivera P, Maccaferri M, Jin Y, Ammar K, Badebo A, Salvi S, Noli E, Crossa J, Tuberosa R (2014) Association mapping reveals novel stem rust resistance loci in durum wheat at the seedling stage. Plant Genome 7:1–13
Long Y, Chao WS, Ma G, Xu SS, Qi, L (2017) An innovative SNP genotyping method adapting to multiple platforms and throughputs. Theor Appl Genet 130:597–607
Mago R, Verlin D, Zhang P, Bansal U, Bariana H, Jin Y, Ellis J, Hoxha S, Dundas I (2013) Development of wheat–Aegilops speltoides recombinants and simple PCR-based markers for Sr32 and a new stem rust resistance gene on the 2S#1 chromosome. Theor Appl Genet 126:2943–2955
McIntosh RA, Luig NH, Johnson R, Hare RA (1981) Cytogenetical studies in wheat. XI. Sr9g for reaction to Puccinia graminis tritici. Z Pflanzenzüchtg 87:274–289
Mergoum M, Frohberg RC, Stack RW, Rasmussen JB, Friesen TL (2008) Registration of ‘Faller’ spring wheat. J Plant Reg 2:224–229
Newcomb M, Acevedo M, Bockelman HE, Brown-Guedira G, Goates BJ, Jackson EW, Jin Y, Njau P, Rouse MN, Singh D, Wanyera R, Bonman JM (2013) Field resistance to the Ug99 race group of the stem rust pathogen in spring wheat landraces. Plant Dis 97:882–890
Newcomb M, Olivera PD, Rouse MN, Szabo LJ, Johnson J, Gale S, Luster DG, Wanyera R, Macharia G, Bhavani S, Hodson D, Patpour M, Hovmøller MS, Fetch TG, Jin Y (2016) Kenyan isolates of Puccinia graminis f. sp. tritici from 2008 to 2014: Virulence to SrTmp in the Ug99 race group and implications for breeding programs. Phytopathology 106:729–736
Niu Z, Klindworth DL, Friesen TL, Chao S, Jin Y, Cai X, Xu SS (2011) Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187:1011–1021
Pu J, Wang Q, Shen Y, Zhuang L, Li C, Tan M, Bie T, Chu C, Qi Z (2015) Physical mapping of chromosome 4J of Thinopyrum bessarabicum using gamma radiation-induced aberrations. Theor Appl Genet 128:1319–1328
Qi L, Echalier B, Friebe B, Gill BS (2003) Molecular characterization of a set of wheat deletion stocks for use in chromosome bin mapping of ESTs. Funct Integr Genomics 3:39–55
Quirin EA (2010) Characterization and quantitative trait loci (QTL) mapping of Fusarium head blight resistance-related traits in the Japanese wheat landrace, PI 81791 (Sapporo Haru Komugi Jugo). Ph.D dissertation. Univ. of Minnesota, St. Paul, MN, p 167
Rouse MN, Jin Y (2011) Stem rust resistance in A-genome diploid relatives of wheat. Plant Dis 95:941–944
Rouse MN, Olson E, Pumphrey M, Jin Y (2011a) Stem rust resistance in Aegilops tauschii germplasm. Crop Sci 51:2074–2078
Rouse MN, Wanyera R, Njau P, Jin Y (2011b) Sources of resistance to stem rust race Ug99 in spring wheat germplasm. Plant Dis 95:762–766
Rouse MN, Nava IC, Chao S, Anderson JA, Jin Y (2012) Identification of markers linked to the race Ug99 effective stem rust resistance gene Sr28 in wheat (Triticum aestivum L.). Theor Appl Genet 125:877–885
Rouse MN, Nirmala J, Jin Y, Chao S, Fetch TG Jr, Pretorius ZA, Hiebert CW (2014) Characterization of Sr9h, a wheat stem rust resistance allele effective to Ug99. Theor Appl Genet 127:1681–1688
Saini J, Faris JD, Rouse MN, Chao S, McClean P, Xu SS (2016) Mapping of stem rust resistance genes from Triticum turgidum ssp. dicoccum. Plant & animal genome conference XXIV, January 08–13, 2016. San Diego, CA. https://pag.confex.com/pag/xxiv/webprogram/Paper20607.html
Semagn K, Babu R, Hearne S, Olse M (2014) Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol Breed 33:1–14
Sepsi A, Molnár I, Molnár-Láng M (2009) Physical mapping of a 7 A.7D translocation in the wheat–Thinopyrum ponticum partial amphiploid BE-1 using multicolour genomic in situ hybridization and microsatellite marker analysis. Genome 52:748–754
Singh RP, Hodson DP, Jin Y, Huerta-Espino J, Kinyua MG, Wanyera R, Njau P, Ward RW (2006) Current status, likely migration and strategies to mitigate the threat to wheat production from Ug99 (TTKS) of stem rust pathogen. CAB Reviews: Perspectives in Agriculture, Veterinary Science, Nutrition and Natural Resources, 1, No. 054, 13 pp
Singh RP, Hodson DP, Huerta-Espino J, Jin Y, Bhavani S, Njau P, Herrera-Foessel S, Singh PK, Singh S, Govindan V (2011) The emergence of Ug99 races of stem rust fungus is a threat to world wheat production. Ann Rev Phytopathol 49:465–481
Smith PH, Hadfield J, Hart NJ, Koebner RMD, Boyd LA (2007) STS markers for the wheat yellow rust resistance gene Yr5 suggest a NBS-LRR-type resistance gene cluster. Genome 50:259–265
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Song L, Lu Y, Zhang J, Pan C, Yang X, Li X, Liu W, Li L (2016) Physical mapping of Agropyron cristatum chromosome 6P using deletion lines in common wheat background. Theor Appl Genet 129:1023–1034
Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M (2004) Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct Integr Genom 4:12–25
Sourdille P, Gandon B, Chiquet V, Nicot N, Somers D, Murigneux A, Bernard M (2010) Wheat Génoplante SSR mapping data release: a new set of markers and comprehensive genetic and physical mapping data. http://wheat.pw.usda.gov/ggpages/SSRclub/GeneticPhysical/. Accessed 2 Dec 2016
Stakman EC, Stewart DM, Loegering WQ (1962) Identification of physiologic races of Puccinia graminis var. tritici. USDA-ARS E617. Rev. ed. Scientific Journal Series Paper no. 4691. Minnesota Agric. Exp. Stn., St. Paul, MN
Syvänen AC (2001) Accessing genetic variation: genotyping single nucleotide polymorphism. Nat Rev Genet 2:930–942
Taylor S, Scott R, Kurtz R, Fisher C, Patel V, Bizouarn F (2010) A practical guide to high resolution melt analysis genotyping. Bio-Rad Laboratories Inc Bulletin 6004. http://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6004.pdf. Accessed 6 Mar 2017
Torada A, Koike M, Mochida K, Ogihara Y (2006) SSR-based linkage map with new markers using an intraspecific population of common wheat. Theor Appl Genet 112:1042–1051
Tsilo TJ, Jin Y, Anderson JA (2007) Microsatellite markers linked to stem rust resistance allele Sr9a in wheat. Crop Sci 47:2013–2020
Turner MK, DeHaan LR, Jin Y, Anderson JA (2013) Wheatgrass-wheat partial amphiploids as a novel source of stem rust and Fusarium head blight resistance. Crop Sci 53:1994–2005
Visser B, Herselman L, Park RF, Karaoglu H, Bender CM, Pretorius ZA (2011) Characterization of two new Puccinia graminis f. sp. tritici races within the Ug99 lineage in South Africa. Euphytica 179:119–127
Wilkinson PA, Winfield MO, Barker GLA, Allen AM, Burridge A, Coghill JA, Edwards KJ (2012) CerealsDB 2.0: an integrated resource for plant breeders and scientists. BMC Bioinform 13:219
Wu S, Pumphrey M, Bai G (2009) Molecular mapping of stem-rust-resistance gene Sr40 in wheat. Crop Sci 49:1681–1686
Xu SS, Jin Y, Klindworth DL, Wang RR-C, Cai X (2009) Evaluation and characterization of seedling resistances to stem rust Ug99 races in wheat–alien species derivatives. Crop Sci 49:2167–2175
Xue S, Zhang Z, Lin F, Kong Z, Cao Y, Li C, Yi H, Mei M, Zhu H, Wu J, Xu H, Zhao D, Tian D, Zhang C, Ma Z (2008) A high-density intervarietal map of the wheat genome enriched with markers from expressed sequence tags. Theor Appl Genet 117:181–189
Yao Z-J, Lin R-M, Xu S-C, Li Z-F, Wan A-M, Ma Z-Y (2006) The molecular tagging of the yellow rust resistance gene Yr7 in wheat transferred from differential host Lee using microsatellite markers. Scientia Agric Sin 39:1146–1152
You FM, Huo N, Gu YQ, Luo M-C, Ma Y, Hane D, Lazo GR, Dvorak J, Anderson OD (2008) BatchPrimer3: a high throughput web application for PCR and sequencing primer design. BMC Bioinform 9:253
Yu L-X, Barbier H, Rouse MN, Singh S, Singh RP, Bhavani S, Huerta-Espino J, Sorrells ME (2014) A consensus map for Ug99 stem rust resistance loci in wheat. Theor Appl Genet 127:1561–1581
Yu GT, Klindworth DL, Friesen TL, Faris JD, Zhong S, Rasmussen JB, Xu S (2015) Development of a diagnostic marker for stem rust resistance gene Sr47 introgressed from Aegilops speltoides into durum wheat. Theor Appl Genet 128:2367–2374
Zhang P, McIntosh RA, Hoxha S, Dong C (2009) Wheat stripe rust resistance genes Yr5 and Yr7 are allelic. Theor Appl Genet 120:25–29
Zurn JD, Newcomb M, Rouse MN, Jin Y, Chao S, Sthapit J, See DR, Wanyera R, Njau P, Bonman JM, Brueggeman R, Acevedo M (2014) High-density mapping of a resistance gene to Ug99 from the Iranian landrace PI 626573. Mol Breed 34:871–881
Acknowledgements
We thank Drs. Shunwen Lu and Guojia Ma for critically reviewing the manuscript. This research was supported in part by funds to S. S. X. provided through a grant from the Bill & Melinda Gates Foundation to Cornell University for the Borlaug Global Rust Initiative (BGRI) Durable Rust Resistance in Wheat (DRRW) Project and the USDA-ARS CRIS Project No. 3060-520-037-00D. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors have no conflict of interest.
Ethical standards
The experiments were performed in compliment with the current laws of United States of America.
Additional information
Communicated by Diane E. Mather.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2017_2875_MOESM1_ESM.pdf
Supplemental Figure S1. Separation of amplicons from primer Xrwgs38 illustrating amplicons designated as Xrwgs38a and Xrwgs38b. Plants 3 thru 7 were derived from the Joppa-RWG35 population and include three heterozygous (#4, 5, and 6) and two homozygous plants (#3 and 7). Supplemental Figure S2. Detection of polymorphisms between Rusty and Joppa observed in RWG35 and RWG37 when amplified with co-dominant markers Xmag1729 and Xrwgs38. A. Marker Xmag1729 separated with 0.5× TBS buffer for 2.0 h at 40 W of current. RWG35 carries the 433 bp amplicon from Rusty and lacks the 444 bp amplicon from DAS15. A-C. Amplicons designated as Xrwgs38b separated in (B) 0.5× TBS buffer for 1.5 h at 70 W of current, (C) 0.5× TBS buffer for 2.0 h at 40 W of current, and (D) 1.0× TBS buffer at 70 W of current. RWG37 carries amplicons from Rusty and lacks the amplicons from DAS15. Figures B-D illustrate that migration of amplicons from Rusty was highly influenced by electrophoretic conditions. Supplemental Figure S3. Effects of number of PCR cycles on amplicon expression of marker Xwmc41. Supplemental Figure S4. Electrophoretograms of 12 dominant repulsion-phase markers tested on the RWG35, RWG36, and RWG37 populations. Only the first 12 plants of each population are shown. When markers produced multiple polymorphic amplicons, only the size of the smallest amplicon is reported. For an explanation of assigning markers to bins, see Fig. 3. X = a plant exhibiting recombination between Joppa and Rusty chromatin. For an explanation of classifying plants for recombination, see Fig. S5. Supplemental Figure S5. Electrophoretograms of co-dominant markers Xgwm120, Xgwm191, and Xgwm388 tested on the RWG36 population. The polymorphisms observed are between Joppa and Rusty. X = a plant exhibiting a recombination between Joppa and Rusty chromatin. Arrows in lane 3 (Plant 1) show the bands that are diagnostic for recombination. The steps to classify plants for recombination are 1) use the check marker (Xrwgs38a) to classify each plant for the presence of Ae. speltoides (S) or Joppa (J) chromatin, and 2) for the marker being tested, classify each plant for the presence of Rusty (R) or Joppa (J) chromatin. For example, for plants 1–5, marker Xrwgs38a indicates genotypes of SJ, SJ, SS, SS, and SJ, respectively. For marker Xgwm388, amplicons of 176 and 173 bp originate from Rusty and Joppa, respectively; and plants 1–5 have genotypes of RJ, RJ, RJ, RR, and RR, respectively. Therefore, plants 3 and 5 are recombinants (PDF 2413 KB)
122_2017_2875_MOESM2_ESM.docx
Supplemental Table S1. Complete listing of the markers investigated during this study, the reason for inclusion in the study, source of information, and PCR conditions for each primer pair (DOCX 22 KB)
122_2017_2875_MOESM3_ESM.pdf
Supplemental Table S2. Complete classification of all markers and plants in three backcross populations used for mapping. Data are ordered by bin (PDF 149 KB)
Rights and permissions
About this article
Cite this article
Klindworth, D.L., Saini, J., Long, Y. et al. Physical mapping of DNA markers linked to stem rust resistance gene Sr47 in durum wheat. Theor Appl Genet 130, 1135–1154 (2017). https://doi.org/10.1007/s00122-017-2875-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-017-2875-7