Abstract
Background
The most frequent cytogenetic abnormality detected in chronic lymphocytic leukemia (CLL) patients is the presence of a deletion within the chromosome band 13q14. Deletions can be heterogeneous in size, generally encompassing the DLEU1 and DLEU2 genes (minimal deleted region), but at times also including the RB1 gene. The latter, larger type of deletions are associated with worse prognosis.
Genomic instability is a characteristic of most cancers and it has been observed in CLL patients mainly associated with telomere shortening.
Case presentation
Cytogenetic and fluorescence in situ hybridization studies of a CLL patient showed a chromosomal translocation t(12;13)(q15;q14), a mono-allelic 13q14 deletion encompassing both the DLEU and RB1 genes, and genomic instability manifested as chromosomal breaks, telomeric associations, binucleated cells, nucleoplasmic bridges, and micronucleated cells.
In conclusion, our CLL patient showed genomic instability in conjunction with a 13q14 deletion of approximately 2.6 megabase pair involving the DLEU and RB1 genes, as well as other genes with potential for producing genomic instability due to haploinsufficiency.
Similar content being viewed by others
Background
Chronic lymphocytic leukemia (CLL) is a B-cell lymphoproliferative disorder commonly affecting elderly people [1]. The most frequent cytogenetic abnormality detected by interphase fluorescence in situ hybridization (FISH) is the presence of a deletion within the chromosome band 13q14. Deletions can be heterogeneous in size, generally encompassing the DLEU1 and DLEU2 genes (minimal deleted region), but at times also including the RB1 gene. The latter, larger type of deletions are associated with worse prognosis [1,2,3].
Genomic instability is present in most cancers. It is characterized by a high frequency of mutations occurring within the cell genome. Alterations in several pathways involved in detecting and repairing DNA damage, telomere maintenance, and chromosomal mitotic segregation will cause increased frequencies of base pair mutation, microsatellite instability, telomere shortening, and chromosome instability mainly manifested as numerical and structural chromosomal abnormalities, micronuclei, and nucleoplasmic bridges (NPB) [4,5,6,7,8]. Several forms of genomic instability has been observed in CLL patients [9,10,11,12,13,14]. We here report a CLL patient with genomic instability and a large mono-allelic 13q14 deletion encompassing the DLEU1, DLEU2 and RB1 genes.
Case presentation
A 61-year-old male patient with bilateral adenomegaly in the neck showed in his peripheral blood a leukocyte count of 49.1 X109/L, with 90% of lymphocytes. Immunophenotyped cells were positive for CD20, CD5, and CD23 surface antigens; therefore, after being diagnosed with CLL (Rai IV), the hematologist administered chemotherapy consisting of cyclophosphamide, adriamycin, vincristine, and prednisone, but the patient’s disease was refractory to such treatment. Next, the patient was started on fludarabicin and rituximab but an adverse reaction was later reported. Another cycle of treatment with cyclophosphamide and prednisone was administered with no response since leukocytosis remained during the 3 years that preceded his demise.
Cytogenetic studies
Peripheral blood lymphocytes obtained before therapy were cultured in RPMI-1640 medium and stimulated with a mixture of phorbol-12-myristate-13-acetate plus pokeweed mitogen at concentrations previously described [15, 16]. After 72 h of incubation, metaphase cells were obtained from cell cultures harvested by standard methods. Chromosomes were stained following the Giemsa-trypsin banding protocol and analyzed under the microscope. Results were interpreted following the ISCN (2016) recommendations [17].
FISH studies
Three fluorescent in situ hybridization (FISH) analyses were performed separately. In a first analysis, we used a mixture of the dual color 13q14.3-deletion probe (Cytocell, LPH 006), which covers the DLEU1 and DLEU2 genes (labeled in red) and the 13q subtelomere sequence (labeled in green), plus the RB1 (13q14) probe (labeled in green; Kreatech, KI-40001). According to information published by the providers, the red labeled probe targeted to the DLEU genes is conformed of two separated fragments of 215 and 93 kb, which together span a sequence from chr13:49962705 to 50,671,242 (hg38; ~ 700 kb). As for the RB1 (13q14) probe, it covers a continuous sequence approximately from chr13:48062708 to 48,801,516 (hg38; ~ 740 kb). A second FISH examination was performed using the MDM2 Amplification probe (Cytocell, LPS 016). We also performed a third FISH study with the dual color P53/ATM probe (Cytocell, LPH 052). In all these FISH studies, cells were counterstained with 4′,6-diamino-2-phenylindole.
Results
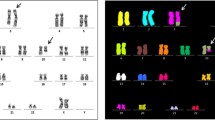
The Giemsa-trypsin banded metaphase analysis displayed the karyotype 46,XY,t(12;13)(q15;q14)[25]/46,XY[2] (Fig. 1 a and b). Seven out of these 25 cells carrying the translocation t(12;13) showed other single-cell abnormalities as chromosomal breaks, translocations, marker chromosomes, and telomeric associations (Fig. 1 a-b). In addition, while performing the chromosomal banding analysis we observed micronucleated and binucleated cells (Fig. 1 c-k). Micronuclei were observed in 68 out of the 1434 scored cells (4.7%), which is within the range of 2.23 to 4.8% of basal micronucleus frequency reported by Hamurcu et al. in six CLL patients [13]. Moreover, thirty out of the 1434 scored cells (2.1%) were binucleated cells; and, eleven of them (0.77%) displayed NPB (Fig. 1 c-g), which is statistically different (p < 0.001; Fisher’s exact test) from the overall baseline NPB frequency reported by Cai et al. in the peripheral blood lymphocytes of 121 healthy individuals from the general population (0.46 ± 0.20 per 1000 binucleated cells) [18].
Cells observed at GTG-banding analysis. a and b Metaphase cells displaying the translocation t(12;13)(q15;q14). Telomeric associations (tas) were also observed in both cells. Moreover, a marker chromosome is shown in b. c-g Binucleated cells showing NPB. h-k Micronucleated cells. Scaling bar = 10 μm
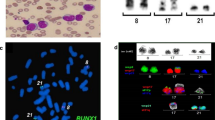
The FISH study with the mixture of DLEU, 13q subtelomere, and RB1 probes aimed to reveal the status of the DLEU and RB1 genes in the derivative chromosomes of the translocation. We found a heterozygous 13q14 deletion of approximately 2.6 megabase pair (nearby from chr13:48062708 to 50,671,242, (hg38)), which included both the DLEU and RB1 genes (Fig. 2 a). Such a deletion was observed in 92% of the 200 scored nuclei. Strikingly, all binucleated cells, as well as cells having micronuclei, analyzed in this FISH experiment, were positive for that deletion (Fig. 2 b-d). Subsequent FISH analysis with the MDM2 probe was done in order to explore the location of the 12q breakpoint in the t(12;13) translocation; such a breakpoint was located centromeric to the MDM2 gene (Fig. 2 e-f). In addition, no evidence of trisomy for chromosome 12 was found after analyzing 200 interphase cells, which consistently showed two MDM2 and two D12Z1 probe signals. Furthermore, FISH study with the dual color P53/ATM probe for searching deletions of these genes disclosed normal results in the 200 scored nuclei (not shown).
FISH observations. a-d FISH study performed with a mixture of DLEU, 13q subtelomere, and RB1 probes. In a, the normal chromosome 13 shows the three expected signals (RB1 (green), DLEU (red) and 13q subtelomere (green)), whereas, only one 13q subtelomere signal is observed on the der(12) chromosome. All analyzed binucleated cells with NPB (b and c), as well as micronucleated cells (d), displayed a signal pattern concordant with RB1-DLEU deletion. e GTG-banded metaphase with translocation t(12;13). f The same metaphase was studied by FISH with the MDM2 amplification probe. The MDM2 red signal is observed on the der(13) chromosome evidencing that the breakpoint on the der(12) occurred centromeric to the MDM2 gene. MN = micronucleus
Discussion and conclusion
The translocation t(12;13)(q15;q14) observed in our patient caused a heterozygous deletion of the DLEU and RB1 genes. There are other translocations registered in the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer [19] that affect the 13q14 chromosomal band. However, there are only four cases of translocation t(12;13) sharing the breakpoints observed in our case [20,21,22,23]. FISH analysis in two of these four cases showed 13q14 deletions of D13S319 or D13S25 markers, which are within sequence of the DLEU1 and DLEU2 genes [22, 23]. The 13q14.3 chromosomal band is the minimal deleted region for B-cell CLL and it is known for its potential tumor-suppressing function. It contains various tumor suppressor gene candidates, mainly the DLEU1 and DLEU2 genes whose loss has been considered as an early step in the development of the disease [24, 25].
It is hard to explain the genomic instability present in our patient since there are several mechanisms that could be related with its presence, as telomere shortening suggested by the presence of telomeric associations, as well as the gene content of the 13q14 region. There are several genes mapping in the deleted segment observed in our patient (chr13:48062708 to 50671242, hg38) whose haploinsufficiency has the potential for causing genome instability. The Mir-16 gene is involved in the DNA damage signaling pathway [26]; the RCBTB2 and SETDB2 genes play a role in chromosome condensation and segregation during mitosis [27, 28]; and, the KPNA3 gene is involved in the nuclear import of protein MeCP2 that has important roles in regulating chromatin structure [29]. Regarding the RB1 gene, its protein plays a well-known role in G1 to S phase progression. Alteration of this mechanism of regulation causes a replicative stress, resulting in the production of DNA double-stranded breaks [30]. Interestingly, new roles of the RB1 protein have been recently identified and all of them are directly involved in the maintenance of genome stability [31, 32]. Coschi et al. [31] linked haploinsufficiency of the RB1 gene with a wide variety of aberrant processes affecting the normal cell cycle as alteration of the number of centrosomes, defects in the mitotic spindle assembly, occurrence of merotelic kinetochore attachments, and failure of cytokinesis generating either binucleated cells or NPB. Therefore, from this perspective, genomic instability could be a variable phenotypic consequence comparable to that occurring in contiguous gene syndromes, where deletions have variable phenotypic manifestations depending mainly on the amount of genetic material lost.
On the other hand, telomeric associations observed in our patient could be true chromosomal translocations rendering dicentric chromosomes and could be interpreted as an indicator of telomere shortening, phenomenon that has been directly associated with genome instability in CLL [9,10,11]. It is well known that during cell division, dicentric chromosomes can be pulled apart towards opposite spindle poles causing an abortion of cytokinesis, which in turn, produces both binucleated cells and NPB [5, 31, 33] as was observed in our patient (Fig. 1 c-g).
In conclusion, our CLL patient showed genomic instability in conjunction with a 13q14 deletion involving the DLEU and RB1 genes. Although studies of CLL patients include the determination of the 13q14 deletion, most of them focus almost exclusively on the DLEU genes. It would be advisable to determine the status of the RB1 gene in those patients with DLEU deletion in order to determine the size of the deletion and to make a better assessment of the prognosis. In addition, more attention should also be paid to the search for biomarkers of genomic instability in patients with CLL, as it could be a more frequent phenomenon than is currently reported.
Abbreviations
- CLL:
-
Chronic lymphocytic leukemia
- FISH:
-
Fluorescent in situ hybridization
- NPB:
-
Nucleoplasmic bridges
References
Kipps TJ, Stevenson FK, Wu CJ, Croce CM, Packham G, Wierda WG, et al. Chronic lymphocytic leukaemia. Nat Rev Dis Primers. 2017;3:16096.
Ouillette P, Collins R, Shakhan S, Li J, Li C, Shedden K, et al. The prognostic significance of various 13q14 deletions in chronic lymphocytic leukemia. Clin Cancer Res. 2011;17:6778–90.
Dal Bo M, Rossi FM, Rossi D, Deambrogi C, Bertoni F, Del Giudice I, et al. 13q14 deletion size and number of deleted cells both influence prognosis in chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2011;50:633–43.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, et al. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011;26:125–32.
Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–45.
Russo A, Pacchierotti F, Cimini D, Ganem NJ, Genescà A, Natarajan AT, et al. Genomic instability: crossing pathways at the origin of structural and numerical chromosome changes. Environ Mol Mutagen. 2015;56:563–80.
Kalsbeek D, Golsteyn RM. G2/M-phase checkpoint adaptation and micronuclei formation as mechanisms that contribute to genomic instability in human cells. Int J Mol Sci. 2017;18.
Véronèse L, Tournilhac O, Callanan M, Prie N, Kwiatkowski F, Combes P, et al. Telomeres and chromosomal instability in chronic lymphocytic leukemia. Leukemia. 2013;27:490–3.
Dos Santos P, Panero J, Palau Nagore V, Stanganelli C, Bezares RF, Slavutsky I. Telomere shortening associated with increased genomic complexity in chronic lymphocytic leukemia. Tumour Biol. 2015;36:8317–24.
Thomay K, Fedder C, Hofmann W, Kreipe H, Stadler M, Titgemeyer J, et al. Telomere shortening, TP53 mutations and deletions in chronic lymphocytic leukemia result in increased chromosomal instability and breakpoint clustering in heterochromatic regions. Ann Hematol. 2017;96:1493–500.
Geyer JT, Subramaniyam S. Micronuclei and nuclear budding in chronic lymphocytic leukaemia. Br J Haematol. 2014;167:585.
Hamurcu Z, Dönmez-Altuntas H, Patiroglu T. Basal level micronucleus frequency in stimulated lymphocytes of untreated patients with leukemia. Cancer Genet Cytogenet. 2008;180:140–4.
Amouroux I, Mossafa H, Gentilhomme O, Girot R, Flandrin G, Troussard X. Chronic lymphocytic leukaemia with binucleated lymphocytes. Leuk Lymphoma. 1997;27:533–7.
Heerema NA, Byrd JC, Cin PD, Dell’ Aquila ML, Koduru P, Aviram A, et al. Stimulation of chronic lymphocytic leukemia (CLL) cells with CpG Oligodeoxynucleotide (ODN) gives consistent Karyotypic results among laboratories: a CLL research consortium (CRC) study. Cancer Genet Cytogenet. 2010;203:134–40.
Muthusamy N, Breidenbach H, Andritsos L, Flynn J, Jones J, Ramanunni A, et al. Enhanced detection of chromosomal abnormalities in chronic lymphocytic leukemia by conventional cytogenetics using CpG oligonucleotide in combination with pokeweed mitogen and Phorbol Myristate acetate. Cancer Genet. 2011;204:77–83.
McGowan-Jordan J, Simons A, Schmid M. ISCN 2016: an International System for Human Cytogenomic Nomenclature. Unionville: S. Karger Publications, Inc; 2016.
Cai TJ, Lu X, Tian XL, Zhao H, Li S, Feng JB, et al. Effects of age and gender on the baseline and 2 Gy 60Co γ-ray-induced nucleoplasmic bridges frequencies in the peripheral blood lymphocytes of Chinese population. Mutat Res. 2018;832–833:29-34.
Mitelman F, Johansson B, Mertens F, editors: Mitelman database of chromosome aberrations and gene fusions in Cancer (2018). https://cgap.nci.nih.gov/Chromosomes/Mitelman. Accessed 14 Jun 2018.
Peterson LC, Lindquist LL, Church S, Kay NE. Frequent clonal abnormalities of chromosome band 13q14 in B-cell chronic lymphocytic leukemia: multiple clones, subclones, and nonclonal alterations in 82 midwestern patients. Genes Chromosomes Cancer. 1992;4:273–80.
Gardiner AC, Corcoran MM, Oscier DG. Cytogenetic, fluorescence in situ hybridization, and clinical evaluation of translocations with concomitant deletion at 13q14 in chronic lymphocytic leukaemia. Genes Chromosomes Cancer. 1997;20:73–81.
Mould S, Gardiner A, Corcoran M, Oscier DG. Trisomy 12 and structural abnormalities of 13q14 occurring in the same clone in chronic lymphocytic leukaemia. Br J Haematol. 1996;92:389–92.
El-Taweel M, Barin C, Cymbalista F, Eclache V. Detection of chromosomal abnormalities associated with chronic lymphocytic leukemia: what is the best method? Cancer Genet Cytogenet. 2009;195:37–42.
Mertens D, Philippen A, Ruppel M, Allegra D, Battacharya N, Tschuch C, et al. Chronic lymphocytic leukemia and 13q14: miRs and more. Leuk Lymphoma. 2009;50:502–5.
Cimmino A, Calin GA, Fabbri M, Lorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9.
Zhang X, Wan G, Mlotshwa S, Vance V, Berger FG, Chen H, et al. Oncogenic Wip1 phosphatase is inhibited by miR-16 in the DNA damage signaling pathway. Cancer Res. 2010;70:7176–86.
Devilder MC, Cadoret E, Chérel M, Moreau I, Rondeau G, Bézieau S, et al. cDNA cloning, gene characterization and 13q14.3 chromosomal assignment of CHC1-L, a chromosome condensation regulator-like guanine nucleotide exchange factor. Genomics. 1998;54:99–106.
Falandry C, Fourel G, Galy V, Ristriani T, Horard B, Bensimon E, et al. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J Biol Chem. 2010;285:20234–41.
Baker SA, Lombardi LM, Zoghbi HY. Karyopherin α 3 and karyopherin α 4 proteins mediate the nuclear import of methyl-CpG binding protein 2. J Biol Chem. 2015;290:22485–93.
Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–46.
Coschi CH, Ishak CA, Gallo D, Marshall A, Talluri S, Wang J, et al. Haploinsufficiency of an RB-E2F1-Condensin II complex leads to aberrant replication and aneuploidy. Cancer Discov. 2014;4:840–3.
Uchida C. Roles of pRB in the regulation of nucleosome and chromatin structures. Biomed Res Int. 2016;2016:5959721.
Gascoigne KE, Cheeseman IM. Induced dicentric chromosome formation promotes genomic rearrangements and tumorigenesis. Chromosom Res. 2013;21:407–18.
Acknowledgements
We thank María de Lourdes Carbajal her review of the writing.
Funding
This work was funded by the “FIS -Instituto Mexicano del Seguro Social”: # FIS/IMSS/PROT/PRIO/16/061. The funding body did not participate either in the design of the study, collection, analysis, or interpretation of data, nor in the writing or reviewing of the manuscript. MPNR received a scholarship from Coordinación de Investigación en Salud, IMSS. México.
Availability of data and materials
The authors declare that all relevant data are included in the article.
Author information
Authors and Affiliations
Contributions
MPNR, MDDC, did laboratory work as karyotyping and FISH, acquired and interpreted data; LBAL and CBG contributed with the diagnosis, management and clinical-hematological follow-up of the patient; MTMT and JRGG interpreted data, designed the figures, and wrote de manuscript. All authors critically read the manuscript. All of them approved and agreed to publish the material contained in this manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The authors declare to have complied with ethical standards. The data presented here are part of a research project approved by our institutional research and ethics committees (Instituto Mexicano del Seguro Social, Project # R-2013-785-071).
Written informed consent was obtained from the patient for participating in this study.
Consent for publication
The patient gave his approval by signing an informed consent.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Nava-Rodríguez, M.P., Domínguez-Cruz, M.D., Aguilar-López, L.B. et al. Genomic instability in a chronic lymphocytic leukemia patient with mono-allelic deletion of the DLEU and RB1 genes. Mol Cytogenet 12, 2 (2019). https://doi.org/10.1186/s13039-019-0417-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-019-0417-5