Abstract
Background
In acute myeloid leukemia (AML), the MDS1 and EVI1 complex locus - MECOM, also known as the ecotropic virus integration site 1 - EVI1, located in band 3q26, can be rearranged with a variety of partner chromosomes and partner genes. Here we report on a 57-year-old female with AML who presented with the rare translocation t(3;10)(q26;q21) involving the MECOM gene. Our aim was to identify the fusion partner on chromosome 10q21 and to characterize the precise nucleotide sequence of the chromosomal breakpoint.
Methods
Cytogenetic and molecular-cytogenetic techniques, chromosome microdissection, next generation sequencing, long-range PCR and direct Sanger sequencing were used to map the chromosomal translocation.
Results
Using a combination of cytogenetic and molecular approaches, we mapped the t(3;10)(q26;q21) to the single nucleotide level, revealing a fusion of the MECOM gene (3q26.2) and C10orf107 (10q21.2).
Conclusions
The approach described here opens up new possibilities in characterizing acquired as well as congenital chromosomal aberrations. In addition, DNA sequences of chromosomal breakpoints may be a useful tool for unique molecular minimal residual disease target identification in acute leukemia patients.
Similar content being viewed by others
Background
EVI1 is one of several protein isoforms encoded by the MECOM locus at human chromosome 3q26 that also yields the MDS1 and MDS1-EVI1 protein isoform[1]. The role of MDS1 and MDS1-EVI1 in malignancy is still unclear, though the EVI1 transcription factor plays an essential role in the proliferation and maintenance of hematopoietic stem cells[2]. Aberrant EVI1 expression occurs in approximately 8% of patients with de novo acute myeloid leukemia (AML)[3]. The overexpression of EVI1 can be achieved not only through rearrangements of band 3q26 but also without the presence of 3q26 abnormalities, therefore indicating that other mechanisms can lead to EVI1 activation[4–6]. Moreover, a substantial number of patients with 3q26 rearrangements do not express EVI1[7]. In approximately 2% of AML cases, inv(3)(q21q26)/t(3;3)(q21;q26) is observed, where it has been suggested that the promoter of the house-keeping RPN1 gene could be responsible for the activation of EVI1[8]. Other EVI1 rearrangements include, e.g. 7q21 (CDK6), 7q34 (TCRB), 12p13 (ETV6) and 21q22 (RUNX1)[6, 9]. Even though partner chromosomes and molecular consequences differ between various types of EVI1 rearrangements, elevated expression predicts poor prognosis for the affected patients[4, 10, 11].
Here we report the rare case of chromosomal translocation t(3;10)(q26;q21) involving MECOM. Using modern cytogenetic and molecular biological techniques we were able to characterize the nucleotide sequence of this breakpoint and thus identify the fusion partner on chromosome 10.
Case presentation
A 57-year old female was diagnosed with AML (FAB M2) after a blood cell count and bone marrow examination was initiated in June 2013. Hematologic parameters were as follows: hemoglobin 6,2 g/dl, platelets 44 × 109/l, and white blood cells (WBC) 3,34 × 109/l with 7,8% neutrophils, 62,9% lymphocytes and 28,7% monocytes, 0% eosinophils and 0,3% basophils. A bone marrow aspirate revealed slightly hypercellular marrow with normocellular particles. Megakaryocytes were found in reduced density. There was significant hiatus leucaemicus with evidence of medium-sized blasts with poor basophilic cytoplasm and distinct granulation. Flow cytometry performed on the bone marrow revealed 31% myeloid-appearing blasts with expression of CD34 and CD117, and confirmed the diagnosis of AML.
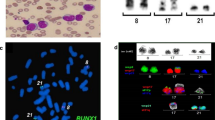
Conventional cytogenetic analysis of a 24-h culture, performed on bone marrow cells by standard techniques and evaluated by G-banding, revealed a balanced t(3;10)(q26;q21) in 20/22 metaphases. Involvement of the MECOM gene was confirmed by FISH with the use of a commercially available probe set.
Results
Cytogenetic and molecular-cytogenetic analyses of bone marrow cultures revealed an aberrant karyotype 46,XX,t(3;10)(q26;q21) – Figure 1. A commercial EVI1 break-apart probe yielded a split signal in all dividing and 80% of the interphase bone marrow cells, demonstrating the rearrangement of the 3q26 chromosomal region (Figure 2).
Ten derivative chromosome 10 breakpoint regions were dissected, amplified and sequenced. In total, 81 753 reads were obtained and aligned to reference sequences of chromosomes 3 and 10 (NCBI build 37.3). Long-range PCR primer design resulted in a product that was then subjected to Sanger sequencing. The nucleotide sequence of the der(10) breakpoint (Figure 3) revealed a fusion of the MECOM gene on 3q26 to C10orf107 on 10q21.
Additionally, the bone marrow sample was subjected to reverse transcription real-time PCR analysis to determine the expression levels of cEVI1 (i.e., the sum of all EVI1 mRNA variants) relative to those of the internal reference gene ABL. We found that EVI1 expression was 26-fold higher when compared with healthy control (data not shown).
Discussion
In the present report we describe a rare case of acute myeloid leukemia with a t(3;10)(q26;q21) translocation involving MECOM. To our knowledge[12], only one case with this translocation has been reported[9], but the fusion partner on chromosome 10 was not characterized. Using a novel technical approach we were able to identify the fusion partner and precise nucleotide sequence of the breakpoint, which may serve as a patient-specific molecular target for subsequent real-time PCR-based minimal residual disease (MRD) monitoring. We further demonstrated by real-time quantitative reverse transcription PCR that the t(3;10)(q26;q21) results in EVI1 over-expression.
Deregulated expression of EVI1 and other genes (e.g. BAALC, WT1) involved in cell proliferation, survival and differentiation have been used as alternative MRD targets[13–16]. However, the sensitivity of expression assays is dependent on the level of initial expression; therefore, these assays are suitable only in AML cases with a high initial expression level of a specific target normalized to an endogenous control gene at diagnosis. Even in those cases, the sensitivity is usually not sufficient for subsequent MRD monitoring. Therefore, in patients presenting with a fusion transcript and/or gene mutation, a specific PCR assay is preferred. These PCR-based methods are currently the most sensitive techniques for MRD follow-up, reaching sensitivities of 10-4 – 10-5[17, 18].
Real-time PCR-based MRD assays allow the highly accurate quantification of residual leukemic cells and evaluations of treatment outcome in AML patients. The merit of MRD monitoring during patient‘s treatment and prognostic relevance has been confirmed by various studies[17, 19, 20]. Common targets for MRD detection include fusion transcripts (e.g. RUNX1-RUNX1T1, PML-RARα, DEK-NUP214, CBFβ-MYH11)[21] and mutations of clinically relevant genes (e.g. NPM1, CEBPα, FLT3, c-KIT)[17–22]. Unfortunately, approximately half of AML patients lack a molecular target suitable for MRD monitoring[23]. Therefore, introducing novel approaches for the identification of unique clone-specific markers is highly desirable. The procedure described here is based on characterizing nucleotide sequences of unique chromosomal breakpoints, allowing the design of a specific real-time PCR assay for MRD assessment. In this way, AML patients could benefit from accurate and sensitive MRD monitoring, even in the absence of other well-introduced molecular marker[24].
Mapping chromosome breakpoints is a conventional method for identifying specific genes in leukemic patients, as well as patients with solid tumors and individuals with balanced translocations[25–27]. A fundamental requirement is the ability to karyotype and precisely identify derivative chromosomes using classic karyotyping or molecular cytogenetic tools such as mFISH and mBAND analyses. Hybridization with even higher resolution, such as BAC-FISH (Bacterial Artificial Chromosome FISH) can help to narrow-down the chromosomal breakpoints further, though it is still not subtle enough to allow subsequent molecular methods to be used and to identify nucleotide sequence. There have been a number of methods proposed to address this issue, with varying strengths and weaknesses. Array-CGH has improved in resolution, allowing deletions, amplifications, and non-balanced translocations to be more precisely characterized, but array-CGH in principle cannot detect targets arising from balanced chromosomal translocations[28].
Conclusion
The combination of cytogenetic and molecular methods described here enabled us to proceed from the chromosomal level (cytogenetically identified abnormality) to the molecular level (unique DNA sequence) in a case of the novel t(3;10)(q26;q21) translocation. Using this procedure, acquired as well as congenital chromosomal aberrations can be characterized. In contrast to other mapping methods (e.g. BAC-FISH, array CGH) our technique allows the rapid mapping of chromosomal breakpoints down to the DNA sequence level and immediate elucidation of possible genes involved. This can be invaluable for studying such aberrations in a wide variety of fields, including the evolution of diseases or the genetic basis of inherited syndromes.
Methods
Cytogenetic and molecular cytogenetic analyses
The heparinized bone marrow sample was cultivated for 24 h in RPMI 1640 media supplemented with 10% fetal calf serum, penicillin/streptomycin and L-glutamine (PAA Laboratories, Austria) at 37°C/5% CO2. Karyotype was investigated by G-banding and multiplex fluorescence in situ hybridization (mFISH) with the 24XCyte probe kit (MetaSystems, Germany). ISCN 2013 nomenclature was used to describe chromosome abnormalities[29]. Interphase fluorescence in situ hybridization (FISH) analysis was performed using a commercially available EVI1 break-apart probe (MetaSystems, Germany).
DNA/RNA isolation, reverse transcription
DNA and RNA were isolated from the mononuclear fraction of bone marrow samples at diagnosis. DNA was isolated using the MagNA Pure automatic isolator (Roche, Germany) according to the manufacturer’s instructions. RNA was extracted by TRI Reagent (Molecular Research Center, USA) according to the manufacturer’s recommendations. Reverse transcription was performed using the Verso cDNA Synthesis Kit (Thermo Scientific, USA) according to the manufacturer’s instructions.
Real-time quantitative reverse transcriptase PCR
Primers and probes to amplify and quantify EVI1-expression were forward: 5′ ACCCACTCCTTTCTTTATGGACC 3′, reverse: 5′ TGATCAGGCAGTTGGAATTGTG 3′, probe: FAM - 5′ TGAGGCCTTCTCCAGGATTCTTGTTTCAC 3′ - BHQ1. Expression was normalized against the expression of the control gene ABL. Primers and probe to quantify ABL gene were as follows: forward: 5′ TCCTCCAGCTGTTATCTGGAAGA 3′, reverse: 5′ TGGGTCCAGCGAGAAGGTT 3′, probe: FAM-5′ CCAGTAGCATCTGACTTTGAGCCTCAGGG 3′ - BHQ1. PCR conditions started with a denaturation at 95°C for 8 minutes, followed by 45 cycles of denaturation at 95°C for 20 s, annealing at 57°C for 30 s and elongation at 72°C for 30 s.
Chromosomal breakpoint identification
The cell suspension and DNA sample were treated and analyzed as previously described[24]. Briefly, regions around the breakpoints of derivative chromosomes were dissected by glass microneedles manipulated by micromanipulator using an inverted microscope (Axiovert 10, Zeiss, Germany). The microdissected fragments were directly subjected to amplification by degenerate oligonucleotide-primed (DOP) PCR and then sequenced on the GS Junior platform (Roche, Germany) for next generation sequencing. Obtained reads were aligned to reference sequences of chromosomes 3 and 10, using in-house developed software. The last mapped reads from both chromosomes were used as docking sites for primers for long-range PCR to amplify the putative breakpoint. Primers for long-range PCR were designed in Vector NTI Advance (v. 11.5, Invitrogen, USA). PCR amplification was done using the Expand Long Range dNTPack kit (Roche, Germany). The long-range PCR product was directly sequenced using Sanger sequencing to reveal the precise nucleotide sequence of the breakpoint.
Consent
Written informed consent was obtained from the patient for publication of this Case Report. A copy of the written consent is available for review by the Editor-in-Chief of this journal.
References
Fears S, Mathieu C, Zeleznik-Le N, Huang S, Rowley JD, Nucifora G: Intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc Natl Acad Sci U S A 1996,93(4):1642–1647.
Goyama S, Kurokawa M: Pathogenetic significance of ecotropic viral integration site-1 in hematological malignancies. Cancer Sci 2009,100(6):990–995.
Nucifora G, Laricchia-Robbio L, Senyuk V: EVI1 and hematopoietic disorders: history and perspectives. Gene 2006, 368: 1–11.
Gröschel S, Lugthart S, Schlenk RF, Valk PJ, Eiwen K, Goudswaard C, van Putten WJ, Kayser S, Verdonck LF, Lübbert M, Ossenkoppele GJ, Germing U, Schmidt-Wolf I, Schlegelberger B, Krauter J, Ganser A, Döhner H, Löwenberg B, Döhner K, Delwel R: High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol 2010,28(12):2101–2107.
Arai S, Yoshimi A, Shimabe M, Ichikawa M, Nakagawa M, Imai Y, Goyama S, Kurokawa M: Evi-1 is a transcriptional target of mixed-lineage leukemia oncoproteins in hematopoietic stem cells. Blood 2011,117(23):6304–6314.
Haferlach C, Bacher U, Grossmann V, Schindela S, Zenger M, Kohlmann A, Kern W, Haferlach T, Schnittger S: Three novel cytogenetically cryptic EVI1 rearrangements associated with increased EVI1 expression and poor prognosis identified in 27 acute myeloid leukemia cases. Genes Chromosomes Canc 2012,51(12):1079–1085.
Langabeer SE, Rogers JR, Harrison G, Wheatley K, Walker H, Bain BJ, Burnett AK, Goldstone AH, Linch DC, Grimwade D, MRC Adult Leukaemia Working Party: EVI1 expression in acute myeloid leukaemia. Br J Haematol 2001,112(1):208–211.
Suzukawa K, Parganas E, Gajjar A, Abe T, Takahashi S, Tani K, Asano S, Asou H, Kamada N, Yokota J, Morishita K, Ihle JN: Identification of a breakpoint cluster region 3′ of the ribophorin I gene at 3q21 associated with the transcriptional activation of the EVI1 gene in acute myelogenous leukemias with inv(3)(q21q26). Blood 1994,84(8):2681–2688.
Lugthart S, Gröschel S, Beverloo HB, Kayser S, Valk PJ, van Zelderen-Bhola SL, Jan Ossenkoppele G, Vellenga E, van den Berg-de RE, Schanz U, Verhoef G, Vandenberghe P, Ferrant A, Köhne CH, Pfreundschuh M, Horst HA, Koller E, von Lilienfeld-Toal M, Bentz M, Ganser A, Schlegelberger B, Jotterand M, Krauter J, Pabst T, Theobald M, Schlenk RF, Delwel R, Döhner K, Löwenberg B, Döhner H: Clinical, molecular, and prognostic significance of WHO type inv(3)(q21q26.2)/t(3;3)(q21;q26.2) and various other 3q abnormalities in acute myeloid leukemia. J Clin Oncol 2010,28(24):3890–3898.
Lugthart S, van Drunen E, van Norden Y, van Hoven A, Erpelinck CA, Valk PJ, Beverloo HB, Löwenberg B, Delwel R: High EVI1 levels predict adverse outcome in acute myeloid leukemia: prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood 2008,111(8):4329–4337.
Kataoka K, Kurokawa M: Ecotropic viral integration site 1, stem cell self-renewal and leukemogenesis. Cancer Sci 2012,103(8):1371–1377.
Mitelman F, Johansson B, Mertens F (Eds): In Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2014. http://cgap.nci.nih.gov/Chromosomes/Mitelman
Weber S, Alpermann T, Dicker F, Jeromin S, Nadarajah N, Eder C, Fasan A, Kohlmann A, Meggendorfer M, Haferlach C, Kern W, Haferlach T, Schnittger S: BAALC expression: a suitable marker for prognostic risk stratification and detection of residual disease in cytogenetically normal acute myeloid leukemia. Blood Cancer J 2014, 4: e173.
Weisser M, Haferlach C, Haferlach T, Schnittger S: Feasibility of using the combined MDS-EVI1/EVI1 gene expression as an alternative molecular marker in acute myeloid leukemia: a report of four cases. Cancer Genet Cytogenet 2007,177(1):64–69.
Cilloni D, Renneville A, Hermitte F, Hills RK, Daly S, Jovanovic JV, Gottardi E, Fava M, Schnittger S, Weiss T, Izzo B, Nomdedeu J, van der Heijden A, van der Reijden BA, Jansen JH, van der Velden VH, Ommen H, Preudhomme C, Saglio G, Grimwade D: Real-time quantitative polymerase chain reaction detection of minimalresidual disease by standardized WT1 assay to enhance risk stratification in acute myeloid leukemia: a European LeukemiaNet study. J Clin Oncol 2009,27(31):5195–5201.
Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, van Putten WL, Valk PJ, van der Poel-van de Luytgaarde S, Hack R, Slater R, Smit EM, Beverloo HB, Verhoef G, Verdonck LF, Ossenkoppele GJ, Sonneveld P, de Greef GE, Löwenberg B, Delwel R: High EVI1 expression predicts poor survival in acute myeloid leukemia: a study of 319 de novo AML patients. Blood 2003,101(3):837–845.
Shook D, Coustan-Smith E, Ribeiro RC, Rubnitz JE, Campana D: Minimal residual disease quantitation in acute myeloid leukemia. Clin Lymphoma Myeloma 2009,9(Suppl 3):S281-S285.
Rossi G, Minervini MM, Carella AM, de Waure C, di Nardo F, Melillo L, D'Arena G, Zini G, Cascavilla N: Comparison between multiparameter flow cytometry and WT1-RNA quantification in monitoring minimal residual disease in acute myeloid leukemia without specific molecular targets. Leuk Res 2012,36(4):401–406.
Abdelhamid E, Preudhomme C, Helevaut N, Nibourel O, Gardin C, Rousselot P, Castaigne S, Gruson B, Berthon C, Soua Z, Renneville A: Minimal residual disease monitoring based on FLT3 internal tandem duplication in adult acute myeloid leukemia. Leuk Res 2012,36(3):316–323.
Perea G, Lasa A, Aventin A, Domingo A, Villamor N, de Llano MPQ, Llorente A, Junca J, Palacios C, Fernandez C, Gallart M, Font L, Tormo M, Florensa L, Bargay J, Martí JM, Vivancos P, Torres P, Berlanga JJ, Badell I, Brunet S, Sierra J, Nomdedeu JF, Grupo Cooperativo para el Estudio y Tratamiento de las Leucemias Agudas y Miel: Prognostic value of minimal residual disease (MRD) in acute myeloid leukemia (AML) with favorable cytogenetics [t(8;21) and inv(16)]. Leukemia 2006,20(1):87–94.
Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, Wheatley K, Harrison CJ, Burnett AK, National Cancer Research Institute Adult Leukaemia Working Group: Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010,116(3):354–365.
Schlenk RF, Döhner K, Krauter J, Fröhling S, Corbacioglu A, Bullinger L, Habdank M, Späth D, Morgan M, Benner A, Schlegelberger B, Heil G, Ganser A, Döhner H, German-Austrian Acute Myeloid Leukemia Study Group: Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med 2008,358(18):1909–1918.
Paietta E: Minimal residual disease in acute myeloid leukemia: coming of age. Hematology Am Soc Hematol Educ Program 2012, 35–42. 10.1182/asheducation-2012.1.35
Jancuskova T, Plachy R, Stika J, Zemankova L, Hardekopf DW, Liehr T, Kosyakova N, Cmejla R, Zejskova L, Kozak T, Zak P, Zavrelova A, Havlikova P, Karas M, Junge A, Ramel C, Pekova S: A method to identify new molecular markers for assessing minimal residual disease in acute leukemia patients. Leuk Res 2013,37(10):1363–1373.
Chen W, Kalscheuer V, Tzschach A, Menzel C, Ullmann R, Schulz MH, Erdogan F, Li N, Kijas Z, Arkesteijn G, Pajares IL, Goetz-Sothmann M, Heinrich U, Rost I, Dufke A, Grasshoff U, Glaeser B, Vingron M, Ropers HH: Mapping translocation breakpoints by next-generation sequencing. Genome Res 2008,18(7):1143–1149.
Lu CM, Kwan J, Baumgartner A, Weier JF, Wang M, Escudero T, Munné S, Zitzelsberger HF, Weier HU: DNA probe pooling for rapid delineation of chromosomal breakpoints. J Histochem Cytochem 2009,57(6):587–597.
De Braekeleer E, Douet-Guilbert N, Basinko A, Morel F, Le Bris MJ, Férec C, De Braekeleer M: Using bacterial artificial chromosomes in leukemia research: the experience at the university cytogenetics laboratory in Brest. France J Biomed Biotechnol 2011, 1: 329471.
Zakaria Z, Ahid MF, Ismail A, Keoh TS, Nor NM, Kamaluddin NR, Esa E, Yuen LK, Rahman EJ, Osman R: Chromosomal Aberrations in ETV6/RUNX1-positive Childhood Acute Lymphoblastic Leukemia using 244 K Oligonucleotide Array Comparative Genomic Hybridization. Mol Cytogenet 2012,5(1):41.
Schaffer LG, McGowan-Jordan J, Schmid M (Eds): ISCN 2013: an international system for human cytogenetic nomenclature. Basel: S. Karger; 2013.
Acknowledgements
The work was supported by Ministry of Industry and Trade of the Czech Republic (Grant FR-TI2/579) and by DAAD.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
TJ, RP, LZ, DWH, LZ participated in the design of the study and carried out molecular cytogenetic and molecular genetic studies; RP designed the computer software and performed the biostatistical analysis; JS carried out the next-generation sequencing; IP, K-AK, AR performed flow cytometry analysis, collected and provided the clinical data; OAKM, NK performed chromosomal microdissection; SP designed and coordinated the study. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Jancuskova, T., Plachy, R., Zemankova, L. et al. Molecular characterization of the rare translocation t(3;10)(q26;q21) in an acute myeloid leukemia patient. Mol Cytogenet 7, 47 (2014). https://doi.org/10.1186/1755-8166-7-47
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1755-8166-7-47