Abstract
Background
Feline and canine babesiosis is an important tick-borne disease caused by parasites of the genus Babesia. The disease has a worldwide distribution and causes serious health problems in domestic and wild canidae and felidae.
Results
Genomic DNA was isolated from blood samples, which were randomly collected from pet dogs (n = 115) and cats (n = 25) in Changsha city of Hunan Province, China. Results of nested PCR assay targeting 18S rRNA gene and partial gene sequencing revealed that seven animals were infected with Babesia species, five dogs (5/115, 4.3%) and two cats (2/25, 8.0%). Sequence analysis showed that four dogs (3.5%) were positive for Babesia canis, and the other one for Babesia vogeli (0.87%). The two cats were infected by Babesia hongkongensis.
Conclusions
The findings of this study will expand knowledge of the distribution of Babesia species and provide important epidemiological information for the control of animal babesiosis in China.
Similar content being viewed by others
Background
Feline and canine babesiosis, an important tick-borne hemoprotozoan disease, is prevalent with a worldwide distribution. This disease is caused by several obligate intracellular parasites of the genus Babesia and is characterized by fever, anemia, icterus, hemoglobinuria/bilirubinuria, anorexia, weight loss, and weakness caused by destruction of erythrocytes [1, 4, 12, 25]. It has a serious impact on health of domestic and wild felids and canids. So far, at least 13 identified Babesia species have been detected in domestic cats or wild felids worldwide, including Babesia felis, Babesia cati, Babesia leo, Babesia hongkongensis, Babesia herpailuri, Babesia pantherae, Babesia lengau, Babesia gibsoni, Babesia canis, Babesia vogeli, Babesia rossi, Babesia presentii, and Babesia microti, together with several unidentified Babesia species [5, 8, 14].
Most studied clinical cases of feline babesiosis are induced by Babesia felis, especially in South Africa. This parasite can infect domestic cats, as well as other felids, such as cheetahs and several wild species [1, 21, 23]. Babesia cati, a less pathogenic Babesia species, is primarily found in India and results in milder clinical disease [1, 12]. Babesia leo was considered to infect lions in South Africa, but it has also been detected in a clinically healthy domestic cat [6, 20]. Babesia hongkongensis infection in cats has only been reported in Hong Kong, China [29]. The clinical course of other Babesia species in felids has not been well characterized [21].
Compared with feline babesiosis, the pathogens, clinical descriptions and geographic distributions of canine babesiosis are well-documented. Three large Babesia species have been identified in dogs, B. canis, B. rossi, and B. vogeli. In addition, an unnamed Babesia species that is closely related to Babesia bigemina, has been described in North Carolina in the United States [4, 11, 12, 22]. Babesia vogeli is widespread in the tropical, subtropical and temperate areas of the world. Babesia canis and B. rossi are distributed mainly in Europe and Southern Africa, respectively [12]. There are three small Babesia species that can infect canids [16]. Babesia gibsoni has a worldwide distribution and is a major pathogen of canine babesiosis [32]. Babesia conradae has not yet been reported outside California [15]; Babesia vulpes has been reported in canids in Spain, Italy, Portugal, Croatia, Germany, and the USA [2, 3, 7].
Since 1985, when Lv et al. diagnosed two cases of canine babesiosis caused by B. gibsoni in China [28], this disease has been reported in several provinces across China, including Jiangsu [17], Jiangxi [33], Zhejiang, Anhui, Henan, Shaanxi, Liaoning, Shandong, and Qinghai [24, 26]. These reports have revealed that B. gibsoni, B. vogeli, and B. canis are the causative agents of canine babesiosis in those investigated regions. For feline babesiosis, there is a lack of case reports and pathogen detection in mainland China. However, limited information is available on the prevalence of canine and feline Babesia in Hunan Province of China. In the present study, the status of Babesia infection was investigated using 115 blood samples from pet dogs and 25 from cats located in Changsha city of Hunan Province, China.
Results
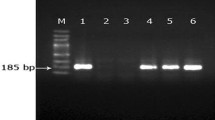
Results of the nested PCR revealed that the percentage of blood samples containing piroplasms was 4.2% (5/115) in domestic dogs and 8.0% (2/25) in cats. To classify the Babesia species detected in this study, long fragments of the 18S rRNA gene (approximately 1400 bp) were successfully amplified from each positive sample for Babesia (Table 1). Results of the sequence analysis were identical to those for the short sequences. The sequences (MH143390–MH143393) shared 99.4–99.9% identity with B. canis derived from dogs in Croatia (AY072926) and Estonia (KT008057). Sequences of MH143394 determined in a poodle dog shared 99.6–99.9% identity with B. vogeli (KY290979, AY072925, AY371198). The two isolates (MH143396, MH143397) from cats were closely related to B. hongkongensis (JQ867356), with 99.6–99.9% sequence identity. Finally, a phylogenetic tree was constructed using the neighbor joining method of MEGA7. The result indicated that the 18S rRNA gene sequences obtained in this study and previously deposited in GenBank were divided into six groups: B. canis, B. vogeli, B. rossi, B. conradae, B. gibsoni and B. hongkongensis (Fig. 1). These data indicate that three Babesia spp. infective to dogs or cats were identified in this study.
Phylogenetic tree based on the sequences of the Babesia 18S rRNA gene (approximately 1400 bp). The tree was constructed using the neighbor joining method of MEGA7 software, and values are given at the nodes. Numbers above the branches demonstrate bootstrap support from 1000 replications. The newly generated sequences in the present study are indicated by bold triangles
Discussion
The prevalence of several tick-borne diseases, caused by Anaplasma phagocytophilum, Rickettsia helvetica, Borrelia burgdorferi and B. canis, is closely related to the spatial and temporal distribution of competent vector ticks in specific regions [10, 13]. The majority of investigations of canine piroplasmosis have been performed during the spring or autumn, the seasons of highest vector tick activity. It has been previously reported in China that the prevalence of B. gibsoni and B. vogeli is 1.5–11.86% and 1.2–11.3%, respectively [9, 18, 30, 33]. In 2019, 130 blood samples were randomly collected from pet dogs in Henan Province to investigate the infection status of piroplasms during the winter. Babesia canis infective to dogs was identified for the first time in China, with a prevalence of 8.8% [26].
In this study, to evaluate the infection status of piroplasms, a small scale investigation using a nested PCR assay combined with gene sequencing was performed in companion animals in Hunan Province. The prevalence of Babesia spp. infections in dogs and cats were 4.3 and 8.0%, respectively, which indicated that there is a prevalence of canine and feline Babesia infection in this area. On the basis of sequence alignment, two canine Babesia species (B. canis, B. vogeli) and one feline Babesia species (B. hongkongensis) were identified in the dog and cat samples, respectively. No B. gibsoni infection was identified in these dogs, but one dog (0.87%) was infected by B. vogeli.
The fact that the pet dogs studied have never travelled outside Changsha city suggested that these parasite infections were locally acquired and endemic in this area. A novel Babesia species was identified in a free-roaming cat and named B. hongkongensis which was previously descried as a feline Babesia in China by Wong et al. [29]. In this study, this species was also identified in two cat samples (8.0%). However, the vector tick of B. hongkongensis is unknown, and this needs to be investigated in the future.
Conclusion
This study has demonstrated a prevalence of Babesia infection in pet dogs and cats. The three previously recorded canine and feline Babesia species (B. vogeli and B. canis and B. hongkongensis) are also prevalent in Hunan Province, China. These data provide valuable information on the distribution of canine and feline Babesia species in China.
Methods
Sample collection and DNA extraction
Between October 2017 and May 2018, 140 blood samples were randomly collected from pet animals, including 115 dogs (61 males and 54 females, 2 months to 16 years old) and 25 cats (nine males and 14 female, 2 months to 5 years old) in animal hospital located in Changsha in the Hunan Province, China. Blood samples were collected in EDTA-coated vacutainer tubes and transported to the laboratory in iceboxes. Genomic DNA was extracted from 200 μL of each blood sample using a commercial DNA extraction kit according to the manufacturer’s instructions (Qiagen DNA blood mini-kit, Germany).
Nested PCR for detection of piroplasms infection
A nested PCR (nPCR) that is universal for piroplasms was used to detect piroplasms infective to dogs and cats as previously reported [26, 27, 31]. Briefly, a set of primers (Piro1-S: 5′-CTTGACGGTAGGGTATTGGC-3′, Piro3-AS: 5′-CCTTCCTTTAAGTGATAAG GTTCAC-3′) was applied to amplify large size fragments of 18S rRNA in the first-round PCR [31]. Moreover, genomic DNA of Babesia bovis and distilled water were used as positive control and the negative control, respectively. Furthermore, small size fragments were amplified using primers (Piro-A: 5′-TTAAATACGAATGCCCCCAAC-3′ and Piro-B: 5′- ATTACCCAATMCBGACACVGKG-3′) [18, 19, 26, 27, 31]. Positive amplicons were purified using a gel DNA purification kit (Zymo, USA), cloned into the pGEM-T Easy vector (Promega, USA). For each amplicon, three positive clones were sequenced using BigDye Terminator Mix (Genscript, Nanjing, China).
PCR amplification of Babesia 18S rRNA gene fragments
To identify the species of piroplasm infecting pet animals, long fragments of the 18S rRNA gene were amplified from positive samples using a nested PCR assay [26, 27]. For each amplicon, three positive clones were sequenced using the BigDye Terminator Mix (Tsingke Biological Technology, China).
Sequences analysis
The 18S rRNA gene sequences obtained in this study were subjected to blast analysis on the NCBI website using the BLASTn program. Representative sequences were deposited in the GenBank database.
A phylogenetic tree was inferred by neighbor joining method using MEGA 7.0 software based on the sequences obtained in this study and the 18S rRNA gene sequences of Babesia spp. previously submitted to GenBank.
Availability of data and materials
DNA sequences obtained in this study have been submitted to GenBank database (accession number: MH143375-MH143379, MH143381-MH143382, MH143390-MH143394, MH143396-MH143397).
Abbreviations
- PCR:
-
Polymerase chain reaction assay
- rRNA:
-
Ribosomal RNA
- EDTA:
-
Ethylene Diamine Tetraacetic Acid
- NCBI:
-
National Center for Biotechnology Information
- bp:
-
Base pair
References
Ayoob AL, Prittie J, Hackner SG. Feline babesiosis. J Vet Emerg Crit Care (San Antonio). 2010;20(1):90–7.
Baneth G, Cardoso L, Brilhante-Simoes P, Schnittger L. Establishment of Babesia vulpes n. sp. (Apicomplexa: Babesiidae), a piroplasmid species pathogenic for domestic dogs. Parasit Vectors. 2019;12(1):129.
Baneth G, Florin-Christensen M, Cardoso L, Schnittger L. Reclassification of Theileria annae as Babesia vulpes sp. nov. Parasit Vectors. 2015;8:207.
Boozer AL, Macintire DK. Canine babesiosis. Vet Clin North Am Small Anim Pract. 2003;33(4):885–904 viii.
Bosman AM, Oosthuizen MC, Peirce MA, Venter EH, Penzhorn BL. Babesia lengau sp. nov., a novel Babesia species in cheetah (Acinonyx jubatus, Schreber, 1775) populations in South Africa. J Clin Microbiol. 2010;48(8):2703–8.
Bosman AM, Venter EH, Penzhorn BL. Occurrence of Babesia felis and Babesia leo in various wild felid species and domestic cats in southern Africa, based on reverse line blot analysis. Vet Parasitol. 2007;144(1–2):33–8.
Harris DJ. Naming no names: comments on the taxonomy of small piroplasmids in canids. Parasit Vectors. 2016;9(1):289.
Hartmann K, Addie D, Belak S, Boucraut-Baralon C, Egberink H, Frymus T, Gruffydd-Jones T, Hosie MJ, Lloret A, Lutz H, Marsilio F, Mostl K, Pennisi MG, Radford AD, Thiry E, Truyen U, Horzinek MC. Babesiosis in cats: ABCD guidelines on prevention and management. J Feline Med Surg. 2013;15(7):643–6.
He L, Miao X, Hu J, Huang Y, He P, He J, Yu L, Malobi N, Shi L, Zhao J. First molecular detection of Babesia gibsoni in dogs from Wuhan, China. Front Microbiol. 2017;8:1577.
Hornok S, Kartali K, Takacs N, Hofmann-Lehmann R. Uneven seasonal distribution of Babesia canis and its two 18S rDNA genotypes in questing Dermacentor reticulatus ticks in urban habitats. Ticks Tick Borne Dis. 2016;7(5):694–7.
Irwin PJ. Canine babesiosis: from molecular taxonomy to control. Parasit Vectors. 2009;2(Suppl 1):S4.
Irwin PJ. Canine babesiosis. Vet Clin North Am Small Anim Pract. 2010;40(6):1141–56.
Kantso B, Svendsen CB, Jensen PM, Vennestrom J, Krogfelt KA. Seasonal and habitat variation in the prevalence of rickettsia helvetica in Ixodes ricinus ticks from Denmark. Ticks Tick Borne Dis. 2010;1(2):101–3.
Kelly PJ, Koster L, Li J, Zhang J, Huang K, Branford GC, Marchi S, Vandenplas M, Wang C. Survey of vector-borne agents in feral cats and first report of Babesia gibsoni in cats on St Kitts, West Indies. BMC Vet Res. 2017;13(1):331.
Kjemtrup AM, Conrad PA. A review of the small canine piroplasms from California: Babesia conradae in the literature. Vet Parasitol. 2006;138(1–2):112–7.
Kjemtrup AM, Wainwright K, Miller M, Penzhorn BL, Carreno RA. Babesia conradae, sp. Nov., a small canine Babesia identified in California. Vet Parasitol. 2006;138(1–2):103–11.
Lanmei Jin QW, Dong Y, Ge L. Investigation on epidemic disease of cannie Babesia gibsoni in Nanjing. J Jinling Inst Technol. 2005;21(4):93–6.
Niu Q, Yang J, Liu Z, Gao S, Pan Y, Guan G, Chu Y, Liu G, Luo J, Yin H. First molecular detection of Piroplasm infection in pet dogs from Gansu, China. Front Microbiol. 2017;8:1029.
Olmeda AS, Armstrong PM, Rosenthal BM, Valladares B, del Castillo A, de Armas F, Miguelez M, Gonzalez A, Rodriguez Rodriguez JA, Spielman A, Telford SR 3rd. A subtropical case of human babesiosis. Acta Trop. 1997;67(3):229–34.
Penzhorn BL, Kjemtrup AM, Lopez-Rebollar LM, Conrad PA. Babesia leo n. sp. from lions in the Kruger National Park, South Africa, and its relation to other small piroplasms. J Parasitol. 2001;87(3):681–5.
Penzhorn BL, Schoeman T, Jacobson LS. Feline babesiosis in South Africa: a review. Ann N Y Acad Sci. 2004;1026:183–6.
Schnittger L, Rodriguez AE, Florin-Christensen M, Morrison DA. Babesia: a world emerging. Infect Genet Evol. 2012;12(8):1788–809.
Schoeman T, Lobetti RG, Jacobson LS, Penzhorn BL. Feline babesiosis: signalment, clinical pathology and concurrent infections. J S Afr Vet Assoc. 2001;72(1):4–11.
Shen Y, Gao J, Xu K, Xue L, Zhang Y, Shi B, Li D, Wei X, Higuchi S. Babesiasis in Nanjing area, China. Trop Anim Health Prod. 1997;29(4 Suppl):19S–22S.
Solano-Gallego L, Baneth G. Babesiosis in dogs and cats--expanding parasitological and clinical spectra. Vet Parasitol. 2011;181(1):48–60.
Wang J, Liu J, Yang J, Liu Z, Wang X, Li Y, Luo J, Guan G, Yin H. Molecular detection and genetic diversity of Babesia canis canis in pet dogs in Henan Province, China. Parasitol Int. 2019a;71:37–40.
Wang J, Liu J, Yang J, Wang X, Li Z, Jianlin X, Li X, Xiang Q, Li Y, Liu Z, Luo J, Guan G, Yin H. The first molecular detection and genetic diversity of Babesia caballi and Theileria equi in horses of Gansu province, China. Ticks Tick Borne Dis. 2019b;10:528–32.
Wenshun Lv XL. The Frist Disceibtion of Babasia gibsoni in China. Chin J Vet Med. 1989;15(12):20–1.
Wong SS, Poon RW, Hui JJ, Yuen KY. Detection of Babesia hongkongensis sp. nov. in a free-roaming Felis catus cat in Hong Kong. J Clin Microbiol. 2012;50(8):2799–803.
Xu D, Zhang J, Shi Z, Song C, Zheng X, Zhang Y, Hao Y, Dong H, Wei L, El-Mahallawy HS, Kelly P, Xiong W, Wang H, Li J, Zhang X, Gu J, Wang C. Molecular detection of vector-borne agents in dogs from ten provinces of China. Parasit Vectors. 2015;8:501.
Yang JF, Li YQ, Liu ZJ, Liu JL, Guan GQ, Chen Z, Luo JX, Wang XL, Yin H. Molecular evidence for piroplasms in wild Reeves’ muntjac (Muntiacus reevesi) in China. Parasitol Int. 2014;63(5):713–6.
Zahler M, Rinder H, Zweygarth E, Fukata T, Maede Y, Schein E, Gothe R. ‘Babesia gibsoni’ of dogs from North America and Asia belong to different species. Parasitology. 2000;120(Pt 4):365–9.
Zheng W, Liu M, Moumouni PF, Liu X, Efstratiou A, Liu Z, Liu Y, Tao H, Guo H, Wang G, Gao Y, Li Z, Ringo AE, Jirapattharasate C, Chen H, Xuan X. First molecular detection of tick-borne pathogens in dogs from Jiangxi, China. J Vet Med Sci. 2017;79(2):248–54.
Acknowledgments
Not applicable.
Funding
This study was financially supported by the National Key Research and Development Program of China (2016YFC1202000); the National Key R&D Program of China (2017YFD0501200); the ASTIP (CAAS-ASTIP-2016-LVRI); the 973 Program (2015CB150300); and the Jiangsu Co-innovation Center Program for Prevention and Control of Important Animal Infectious Disease and Zoonoses. The funding bodies play roles in sample collection, regents, molecular assays and manuscript writing.
Author information
Authors and Affiliations
Contributions
JW wrote the draft of the manuscript. GG, JL and HY designed the study and corrected the manuscript. JW, XW, HS, ZL and YL collected samples and carried out the molecular assays, respectively. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The present study was approved by the Animal Ethics Committee of the Lanzhou Veterinary Research Institute, Academy of Agricultural Sciences (CAAS) (Permit No. LVRIAEC-2018-001). All the procedures were conducted according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China.
Each of the pets’ owners wrote consent and consented to this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, J., Wang, X., Sun, H. et al. Molecular evidence of piroplasm infection in companion animals in Hunan Province, China. BMC Vet Res 16, 297 (2020). https://doi.org/10.1186/s12917-020-02500-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12917-020-02500-6