Abstract
Background
The emergence and spread of multidrug-resistant gram-negative bacteria (MDR) has become a major public health concern worldwide. This resistance is caused by enzymes-mediated genes (i.e., extended spectrum beta-lactamases) that are common in certain Enterobacterioceae species. However, the distribution of these genes is poorly documented in Burkina Faso. This study aims to determine the prevalence and distribution of the resistant genes coding for broad spectrum beta-lactamases and quinolones in rural Burkina Faso.
Methods
Multiplex PCR assays were carried out to detect ESBL-encoding genes, including blaOXA, blaTEM, blaCTX-M, blaSHV. The assays also assessed the presence of quinolone resistance gene namely qnrA, qnrB and qnrS in the quinolone-resistance DEC and Salmonella strains.
Results
The Extended-Spectrum Beta-Lactamases (ESBL) resistance phenotype was reported in all the E. coli isolates (5/5). Cross-resistance phenotype to quinolones (CRQ) was shown by one Salmonella strain (1/9) and three E. coli (3/5). Cross-resistance phenotypes to fluoroquinolones (CRFQ) were harboured by one Salmonella (1/9) and carbapenemase phenotypes were detected in two E. coli strains (2/5). Whilst the blaOXA genes were detected in 100% (5/5) of E. coli isolates and in 33.33% (3/9) Salmonella isolates. One strain of E. coli (1/5) harbored the blaCTX−M gene and the qnrB gene simultaneously.
Conclusions
This study identified β-lactam (bla) and quinolone resistance (qnr) genes in multidrug-resistant E. coli and Salmonella spp. in rural Burkina Faso. Our finding which highlighted the enterobacteriaceae strains resistance to β-lactams and quinolones are of high interest for adequate management of antimicrobial resistant genes outbreak in Burkina Faso.
Similar content being viewed by others
Background
Diarrheal disease is the second leading cause of death among children aged below 5 years [1]. These diseases are especially common in developing countries with poor hygiene and sanitation and with limited access to safe drinking water [2, 3]. In our previous study, the overall prevalence of gastrointestinal infections was 19.7% in children group [4]. These infections are due to bacteria such as Escherichia coli and Salmonella which remain major contributors to acute enteric infection in children. However, the emergence and spread of multidrug-resistant gram-negative bacteria (MDR) has become a major public health concern worldwide [5]. Extended-spectrum beta-lactamases (ESBL) producing Enterobacteriaceae isolates, particularly in Escherichia coli, have been frequently reported in recent years at global scale [6,7,8]. Indeed, ESBL- producing Enterobacteriaceae (ESBL-PE) are associated with high morbidity and mortality rates, prolonged hospital stays and increased costs of healthcare[9, 10]. Some studies have shown that ESBL are responsible for producing antibiotic-resistant bacteria strains [11, 12]. The spread of the strains is likely to limit the effectiveness of antimicrobials used to treat the patients suffering from pathogen bacteria such as Escherichia coli and Salmonella [13,14,15,16]. These ESBL-producing bacteria often show resistance to several antimicrobials such as third and fourth generation cephalosporins as well as quinolones and aminoglycosides [17,18,19]. Although inhibited by clavulanic acid, ESBL enzymes have the ability to hydrolyze third generation cephalosporins and aztreonam [9].
The first ESBL strain which was a Klebsiella ozaenae resistant to oxyimino-cephalosporins was discovered in Germany [20]. In addition to β-lactams, fluoroquinolone resistance due to Qnr genes is emerging and this may pose a challenge in treatment of typhoid in future. These genes belong to the family of repeat pentapeptides that are capable of binding to DNA gyrase and topoisomerase IV, and thus protecting them from inhibitory activities of quinolones [21]. The resistance to quinolones (qnrA) mediated by plasmids in an isolate of Klebsiella pneumoniae was first reported in 1998 from the United States [22]. The excessive use of antibiotics, in particular β-lactams, leads to the selection of ESBL producing strains [23]. However, in developing countries, E. coli identification and microbial drug resistance tests have been limited by phenotypic methods.
Although several antibiotic resistance gene studies have been carried out in Burkina Faso, these studies have been solely conducted in Ouagadougou and Bobo-Dioulasso’s hospitals [24,25,26]. Therefore, the objective of the present study was to determine the prevalence and distribution of resistance gene coding for broad spectrum beta-lactamases and quinolones in two remote rural health centres (Boromo and Gourcy). The main economic activities in these communities are subsistence farming, animal husbandry and commercial activities [4]. Abuse of antibiotics use in animal husbandry has been highlighted in the country with prevalences of antimicrobial residues of 31% and 51.72% in meat and raw milk, respectively [27, 28]. Although antimicrobial use for animals is under veterinary prescription control in Burkina Faso, farmers still use unprescribed antimicrobials as growth promoters or treatment for cattle, poultry and swine [29, 30]. However, there are similarities between the antibiotics employed in agriculture and veterinary and those prescribed for humans in terms of types and mode of actions [31]. Therefore, consumption of contaminated animal-derived food products by residual antibiotics may pose serious public health concerns. In rural settings of Burkina Faso and in many parts of Africa where there is no enough healthcare facilities [32, 33], a high rate of ESBL producing Enterobacteriaceae contaminations is expected among children which might lead to high infantine mortality rates.
Methods
Bacterial isolates
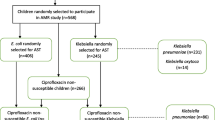
Strains were obtained from our previous studies [34, 35] conducted in Gourcy and Boromo hospitals’ (Fig. 1). 16-plex PCR was used to detect simultaneously 16 genes from the five main pathogroups of E. coli (enterohemoragic E. coli: EHEC, enteropathogenic E. coli: EPEC, enteroaggregative E. coli: EAEC, enteroinvasive E. coli: EIEC and enterotoxigenic E. coli: ETEC) [36]. Furthermore, all Salmonella isolates were serotyped with the somatic O and flagellar H anti-sera according to the Kauffman-White scheme [37].
Map of Burkina Faso. In dark = Gourcy and Boromo where the study was conducted. Legend: Software: Quantum GIS (QGIS 2.2), Valmiera, https://qgis.org/downloads/
Antimicrobial susceptibility test and ESBL production
Antibiotic susceptibility was determined on Mueller-Hinton agar using the standard disc diffusion procedure as described by the European Committee of Antimicrobial Susceptibility Testing (EUCAST) [38]. Nineteen antibiotics belonging to 7 different families were tested as shown in Table 1 (Bio-Rad, France). The diameters of the antibiotic sensitivity halos were recorded according to the EUCAST recommendations. Intermediate (I) susceptibility of pathovars was classified as resistant (R). A double synergy test was used for ESBL-producing strains testing. This consisted of placing discs (2–3 cm diameter) of ceftriaxone and cefotaxime around an amoxicillin-clavulanic acid disc on the bacterial plate.
Molecular identification of resistance genes
DNA extraction was performed using heating method [39]. A loopful of bacterial growth from Mueller Hinton agar (Liofilchem, Italy) plate was suspended in 1 ml of sterilized water. The mixture was boiled for 10 min at 100 °C and centrifuged for 10 min at 12,000 rpm at + 4 °C. Supernatant was then collected and used in the PCR reactions as DNA matrices. Multiplex PCR assays were performed to assess ESBL-encoding genes, including blaOXA, blaTEM, blaCTX−M, blaSHV and the presence of quinolone resistance genes including qnrA, qnrB, qnrS from the quinolone-resistant DEC and Salmonella strains. Primers (GeneCust, France) used for these amplifications are described in Table 2. The PCR assays were carried out in a 25 ml reaction mixture, which consisted of 2.5 µl of the supernatant added to 22.5 µl reaction mixture. This mixture contained 5U of Taq DNA polymerase (Accu Power, South Korea), deoxyribonucleic triphosphate (10 mM), buffer GC (10X), MgCl2 (25 mM) and PCR primers (10 µM). Thermocycling conditions were as follows: 5 min at + 94 °C, followed by 35 amplification cycles at + 94 °C for 30 s, + 59 ± 4 °C for 60 s and + 72 °C for 60 s with a final extension of + 72 °C for 10 min on a thermal cycler (AB Applied Biosystems). Following PCR, the reaction products were separated using electrophoresis in 1.5% agarose gel (weight/volume), stained with Redsaf solution (Prolabo, France) and visualized under UV light (Gel Logic 200) [39].
Results
Antimicrobial resistance
At least, 5 diarrheagenic E. coli and 9 Salmonella strains were identified from our previous studies in 2019 and 2018 respectively. The strains of E. coli identified exhibited a strong resistance to beta-lactams with 100% resistant to amoxicillin-clavulanic acid and amoxicillin, 80% resistant to piperacillin, 60% resistant to cefotaxime, ceftriaxone, aztreonam, cefixime, cefepime and piperacillin-tazobactam. These strains were less resistant to quinolones, 60% resistant to nalidixic acid and no resistant to ciprofloxacin (Fig. 2). By contrast, the Salmonella strains exhibited 100 and 89% resistance to amoxicillin and amoxicillin-clavulanic acid, respectively. Likewise, the resistance of Salmonella to cefixime and cefepime, ceftriaxone and cefotaxime were 67 and 56%, respectively. Salmonella isolates harboured low resistance to quinolones (22% to nalidixic acid and 11% to ciprofloxacin) (Fig. 2).
Associated resistance phenotypes
The distribution of the associated resistance phenotypes is shown in Table 3. Extended-Spectrum Beta-Lactamases (ESBL) resistance phenotype was reported in all E. coli isolates (5/5). Cross-resistance phenotype to quinolones (CRQ) was shown by one Salmonella strain (1/9) and three E. coli (3/5). The Cross-resistance phenotypes to fluoroquinolones (CRFQ) were harboured by one Salmonella (1/9) and carbapenemase phenotypes were detected in two E. coli strains.
Characterization of β-lactamase and quinolones genes
Molecular characterization of E. coli and Salmonella isolates revealed that they harboured several ß-lactamase-encoding genes (blaOXA and blaCTX−M). The blaOXA genes were detected in 100% (5/5) of E. coli isolates and in 33.33% (3/9) Salmonella isolates (Fig. 3). The blaCTX−M gene was detected in one strain of E. coli and this strain also harboured the qnrB gene. The qnrA and qnrS genes were not detected in any of E. coli and Salmonella strains. The distribution of the different genes encoded is shown in Table 2. The genes blaTEM, blaSHV, qnrA and qnrS were not found in this study.
Discussion
The current study was undertaken to screen the ESBL and fluoroquinolone resistance genes among E. coli and Salmonella isolated in children with diarrhea in two rural communities of Burkina Faso. Consistent with global reports, an alarming increase in resistance to beta-lactam antibiotics (even to the extended-spectrum subclass) among clinical E. coli isolates is highlighted by the results of this study. Indeed, the proportion of resistant strains was 60% for E. coli and 50% for Salmonella. These findings were consistent with previous studies in developing countries which showed a resistant rate greater than 50% [40]. This high resistance is likely due to the extensive and excessive clinical use of antibiotics.
Our study showed that all the E. coli isolates (5/5) were ESBL producers in agreement with 95.60% reported in Togo [41]. The presence of ESBL-producing bacteria in hospitals is a major challenge that affects both developed and developing countries [26]. It is known that β-Lactams (mainly extended- past spectrum cephalosporins and carbapenems) and fluoroquinolones constitute the main therapeutic choices to treat infections caused by Enterobacteriaceae. Although carbapenems are the most effective against Gram-positive and Gram-negative bacteria presenting a broad spectrum of antibacterial activity [42], our findings revealed a strong resistance to beta-lactams and moderate rates of resistance to quinolones in E. coli and Salmonella isolated. Indeed, Cross-resistance phenotypes to quinolones (CRQ), Cross-resistance phenotypes to fluoroquinolones (CRFQ) and carbapenemase phenotypes were associated with different rates to our Salmonella and E. coli strains. In agreement with our results, resistance to these compounds has been reported increasingly in several countries [11, 43, 44], limiting dramatically treatment options. Therefore, older agents, such aspolymyxins and fosfomycin, which were rarely implemented in the past because of efficacy and/or toxicity concerns, together with the newer tige-cycline, have become last-resort choices [42].
According to the existing data, this study is the first of kind on rural samples of Burkina Faso. However, it has been shown that fecal carriage of ESBL-PE isolates is one of the main drivers for their dissemination in hospital and community settings worldwide [45]. Because of this mode of diffusion, the ESBLs constitute a significant threat for the countries of West Africa where `the weak socio-economic conditions result in poor hygienic conditions, promoting the spread of resistance. Indeed, most of the farms in Burkina Faso are mainly traditional with unhygienic practices [27]. Furtheremore, our previous study reported that the people in households mainly use well water as a source of drinking [34].
The present study showed that the blaOXA genes were the most common ß-lactamase-producing genes (57.14%), followed by blaCTX−M (7.14%). These findings contrast with those previously reported in Burkina Faso [8, 26]. Otherwise, a spread of blaCTX−M, particularly CTX-M-15 in community and hospital settings has been reported [32, 45, 46]. This difference could be explained by the weakness of the number of multiresistant strains of enterobacteria tested in our study. On the other hand, we noted the simultaneous presence of the blaCTX−M and blaOXA genes in the same strain of E. coli. Our finding confirms the frequent association between blaCTX−M−15 and blaOXA−1 genes in ESBL-PE isolates which has been reported [45, 47,48,49]. This coexistence could reduce the therapeutic options for treatment with β-lactam antibiotics. Thus, combined production of CTX-M and OXA enzymes by E. coli and K. pneumoniae improved resistance to b-lactamase inhibitors, presumably explaining their non-susceptibility to amoxicillin/clavulanate [45, 49, 50]. The genes blaTEM and blaSHV were not identified in the present study. In contrast, these genes have been previously reported in three major hospitals of Ouagadougou [26]. A future study based on more multiresistant strains producing ESBL would shed more light on the existence and prevalence of these genes among rural dwellings.
We also reported the prevalence of plasmid-mediated quinolone resistance in Salmonella and E. coli. Only a single isolate of E. coli (20%) was positive for the qnrB gene which is lower than 67.21% reported in Togo [41] and higher than 3.17% reported in Niger [51]. No Salmonella strain was positive for the qnr genes in the present study. In France, a study revealed 0.2% of qnrA in single isolate of Salmonella [52]. These results may indicate a low dissemination rate of qnr genes among human Salmonella and E. coli isolates. Morever, the E. coli strain that harbored qnrB gene was also positive to ESBL and carbapenemase phenotypes. Indeed, qnr are genes that confer resistance to nalidixic acid and reduced susceptibility to fluoroquinolones [53] and there is frequent association of genes coding for expanded-spectrum b-lactamases (ESBLs) and these genes [52].
The main limitation of the present study consists of the low number of isolates which makes generalizability difficult. Further studies consisting of larger sample size than the number of multidrug-resistant isolates considered in the present study would be necessary. Despite this, the results of this study alert us to (i) the emergence and spread of antibiotic resistance in young children, (ii) the existence of bla and qnr genes in rural areas of Burkina Faso. In addition, the absence of these genes in certain investigated strains maybe due to other mechanisms of resistance to beta-lactams and quinolones.
Conclusions
This study characterized some bla and qnr genes circulating in rural settings that are characterized by their easy transfer between bacteria. The results should contribute to the establishment of a surveillance system for antibiotic resistance in Burkina Faso. Indeed, the data gathered is of paramount importance since it may contribute to design strategies to curtail the emergence and spread of ESBL-producing Enterobacteriaceae among children in rural Burkina Faso and devise innovative therapeutic approaches against multidrug-resistant strains. The intestinal carriage of ESBL-PE is a significant challenge for public health, and highlights the urgent necessity to improve sanitation and implement antibiotic stewardship in developing countries.
Availability of data and materials
All data obtained are available within the article.
Abbreviations
- ESBL:
-
Extended-Spectrum Beta-Lactamases
- CRQ:
-
Cross-resistance phenotype to quinolones
- CRFQ:
-
Cross-resistance phenotypes to fluoroquinolones
- ESBL-PE:
-
Extended-Spectrum Beta-Lactamases producing Enterobacteriaceae
- EUCAST:
-
European Committee of Antimicrobial Susceptibility Testing
References
Abebe W, Earsido A, Taye S, Assefa M, Eyasu A, Godebo G. Prevalence and antibiotic susceptibility patterns of Shigella and Salmonella among children aged below five years with Diarrhoea attending Nigist Eleni Mohammed memorial hospital, South Ethiopia. BMC Pediatr. 2018;18:241.
UNICEF/WHO. Diarrhoea: why children are still dying and what can be done. Geneva: WHO Press; 2009. p. 1–68. Available from http://www.who.int/topics/diarrhoea/en.
Bonkoungou IJO, Haukka K, Österblad M, Hakanen AJ, Traoré AS, Barro N, et al. Bacterial and viral etiology of childhood diarrhea in Ouagadougou, Burkina Faso. BMC Pediatr. 2013;13:36.
Dembélé R, Huovinen E, Yelbéogo D, Kuusi M, Sawadogo G, Haukka K, et al. Burden of acute gastrointestinal infections in Ouagadougou, Burkina Faso. J Microbiol Infect Dis. 2016;6(2):45–52.
Dobiasova H, Dolejska M, Jamborova I, Brhelova E, Blazkova L, Papousek I, et al. Extended spectrum beta-lactamase and fluoroquinolone resistance genes and plasmids among Escherichia coli isolates from zoo animals, Czech Republic. FEMS Microbiol Ecol. 2013;85:604–11.
Holstein A, Grillon A, Yzon L, Morange V, Baty G, Lartigue MF, et al. Prevalence of extended-spectrum b-lactamases of the CTX-M type producing Escherichia coli and Klebsiella pneumoniae in Bretonneau hospitals (CHRU tours). Pathol Biol. 2010; 58:67–69.
Dembélé R, Bonkoungou IJO, Konaté A, Bsadjo-Tchamba G, Bawa HI, Bako E, et al. Serotyping and antibiotic resistance of Enteropathogenic Escherichia coli and E. coli O157 isolated from diarrheal children in rural area of Burkina Faso. Afr J Microbiol Res. 2015;9:1053–59.
Ouédraogo AS, Sanou M, Kissou A, Sanou S, Solaré H, Kaboré F, et al. High prevalence of extended-spectrum β-lactamase producing Enterobacteriaceae among clinical isolates in Burkina Faso. BMC Infect Dis. 2016;16:326.
Pana ZD, Zaouti T. Treatment of extended-spectrum β-lactamase-producing (ESBLs) infections: what have we learned Enterobacteriaceae until now? F1000 Res. 2018; 7. doi:https://doi.org/10.12688/f1000research.14822.1.
Diop A, Sambe-Ba B, Seck A, Dia ML, Timbiné LG, Niang AA, et al. First description of the extended spectrum beta-lactamase gene blaCTX-M-109 in salmonella grumpensis strains isolated from neonatal nosocomial infections in Dakar, Senegal. Plos One. 2016;29:e0157683. https://doi.org/10.1371/journal.pone.0157683.
Cantón R, Novais A, Valverde A, Machado E, Peixe L, Baquero F, et al. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect. 2008;14:144–53.
Hawkey PM. Prevalence and clonality of extended-spectrum β-lactamases in Asia. Clin Microbiol Infect. 2008;14:159–65.
Picozzi SCM, Casellato S, Rossini M, Paola G, Tejada M, Costa E, et al. Extended-spectrum beta-lactamase-positive Escherichia coli causing complicated upper urinary tract infection: Urologist should act in time. Urol Ann. 2014;6(2):107–12.
Ranjbar R, Ghazi FM, Farshad S, Giammanco GM, Aleo A, Owlia P, et al. The occurrence of extended-spectrum β-lactamase producing Shigella spp. in Tehran, Iran. Iran J Microbiol. 2013;5(2):108–12.
Ranjbar R, Giammanco GM, Aleo A, Plano MRA, Naghoni A, Owlia P, et al. Characterization of the first extended-spectrum β-lactamase-producing nontyphoidal Salmonella strains isolated in Tehran, Iran. Foodborne Pathog Dis. 2010;7(1):91–5.
Ghafourian S, Bin Sekawi Z, Sadeghifard N, Mohebi R, Neela VK, Maleki A, et al. The prevalence of ESBLs producing Klebsiella pneumoniae isolates in some major hospitals, Iran. Open Microbiol J. 2011;5:91–5.
Nicolas-Chanoine MH, Bertrand X, Madec JY. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev. 2014; 27: 543–4.
Johnson JR, Nicolas-Chanoine MH, DebRoy C, Castanheira M, Robicsek A, Hansen G, et al. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967–2009. Emerg Infect Dis. 2012;18:598–607.
Rehman MA, Yin X, Lepp D, Laing C, Ziebell K, Talbot G, et al. Genomic analysis of third generation cephalosporin resistant Escherichia coli from dairy cow manure. Vet Sci. 2017;4(4):57.
Kliebe C, Nies BA, Meyer JF, Tolxdorff-Neutzling RM, Wiedemann B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985;28:302–7.
Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–40.
Martınez-Martınez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet. 1998;351:797–9.
Chang YT, Coombs G, Ling T, Balaji V, Rodrigues C, Mikamo H, et al. Epidemiology and trends in the antibiotic susceptibilities of Gram-negative bacilli isolated from patients with intra-abdominal infections in the Asia-Pacific region, 2010–2013. Int J Antimicrob Agents. 2017;49:734–9.
Mètuor-Dabiré A, Zongo KJ, Zèba B, Traoré-Ouedraogo Rasmata M, Jihad B, Marie, et al. First detection of shv-type extended spectrum β-lactamases in the University Hospital complex Paediatric Charles De Gaulle (CHUP-CDG) of Ouagadougou in Burkina Faso. J Asian Sci Res. 2014;4:214–21.
Ouédraogo A-S, Sanou S, Kissou A, Poda A, Aberkane S, Bouzinbi N, et al. Fecal Carriage of Enterobacteriaceae Producing Extended-Spectrum Beta-Lactamases in Hospitalized Patients and Healthy Community Volunteers in Burkina Faso. Microbial Drug Resist. 2017;23:1.
Kpoda DS, Ajayi A, Somda M, Traore O, Guessennd N, Ouattara AS, et al. Distribution of resistance genes encoding ESBLs in Enterobacteriaceae isolated from biological samples in health centers in Ouagadougou, Burkina Faso. BMC Res Notes. 2018;11:471.
Bagré TS, Samandoulougou S, Traoré M, Illy D, Bsadjo-Tchamba G, Bawa-Ibrahim H, et al. Détection biologique des résidus d’antibiotiques dans le lait et produits laitiers de vache consommés à Ouagadougou, Burkina Faso. J Appl Biosc. 2015;87:8105–12.
Samandoulougou S, André JI, Bagre TS, Tapsoba FW, Savadogo A, Scippo M-L, et al. Screening of antibiotics residues in beef consumed in Ouagadougou, Burkina Faso. Afr J Food Sci. 2015;9(6):367–71.
Kagambèga A, Lienemann T, Aulu L, Traoré AS, Barro N, Siitonen A, et al. Prevalence and characterization of Salmonella enterica from the feces of cattle, poultry, swine and hedgehogs in Burkina Faso and their comparison to human Salmonella isolates. BMC Microbiol. 2013;13:253.
Samandoulougou S, Ilboudo AJ, Sanon/ouédraogo G, Tindano K, Compaore H, Ouedraogo A, et al. Utilisation de médicaments vétérinaires en production bovine et aviaire et sécurité sanitaire des aliments d’origine animale au Burkina Faso. Int J Mult Disc Sci. 2016;1(4):1–11.
Saiful Islam KBM, Shiraj-Um-Mahmuda S, Hazzaz-Bin-Kabir M. Antibiotic usage patterns in selected broiler farms of Bangladesh and their public health implications. J Public Health Dev Ctries. 2016;2:276–84.
Farra A, Frank T, Tondeur L, Bata P, Gody JC, Onambele M, et al. High rate of faecal carriage of extended-spectrum b-lactamase-producing Enterobacteriaceae in healthy children in Bangui, Central African Republic. Clin Microbiol Infect. 2016;22(10):891. e1-891.e4.
Tellevik MG, Blomberg B, Kommedal Ø, Maselle SY, Langeland N, Moyo SJ. High prevalence of faecal carriage of ESBL-Producing Enterobacteriaceae among children in Dar es Salaam, Tanzania. PLoS One. 2016;11(12):e0168024.
Dembélé R, Konaté A, Soulama I, Kagambèga A, Kaboré WAD, Cissé H, et al. Prevalence of Multidrug-resistant Salmonella enterica and associated factors among under five children with diarrhea in rural Burkina Faso. Clin Biotechnol Microbiol. 2018;3(1):566–76.
Dembélé R, Konaté A, Kagambèga A, Soulama I, Kaboré WAD, Traoré O, et al. Class 1 Integrons, Genetic Factor for the Dissemination of Tetracycline and Chloramphenicol Resistance Genes in Escherichia coli isolated from Children with Diarrhea in Rural Burkina Faso. EC Microbiol. 2019;15(6):463–70.
Antikainen J, Tarkka E, Haukka K, Siitonen A, Vaara M, Kirveskari J. New 16 plex PCR method for rapid detection of diarrheagenic Escherichia coli directly from stool samples. Europ J Clin Microbiol Infect Dis. 2009;28(8):899–908.
Popoff MY, Bockemuhl J, Gheesling LL. Supplement. 2002 (no. 46) to the Kauffmann-White scheme. Res Microbiol. 2004;155:568–70.
European Committee on Antimicrobial Susceptibility Testing (EUCAST). Recommandation 2017. Éd. V.1.0 Mars. 1–127.
Moyo SJ, Maselle SY, Matee MI, Langeland N, Mylvaganam H. Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC Infect Dis. 2007;7:1–7.
Wang G, Clark CG, Rodgers FG. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J Clin Microbiol. 2002;40:3613–19.
Salah FD, Soubeiga ST, Ouattara AK, Sadji AY, Metuor-Dabire A, Obiri-Yeboah D, et al. Distribution of quinolone resistance gene (qnr) inESBL-producing Escherichia coli and Klebsiella spp. in Lomé, Togo. Antimicrob Resist Infect Control. 2019;8:104.
Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 2016;3(1):15–21.
Ben-Ami R, Rodrıguez-Bano J, Arslan H, Pitout JD, Quentin C, Calbo ES, et al. A multinational survey of risk factors for infection with extended-spectrum betalactamase-producing Enterobacteriaceae in nonhospitalized patients. Clin Infect Dis. 2009;49:682–90.
Johnson JR, Urban C, Weissman SJ, Jorgensen JH, Lewis JS 2nd, Hansen G, et al. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25: H4) and blaCTX-M-15 among extended-spectrum-b-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob Agents Chemother. 2012;56:2364–70.
Ouchar Mahamat O, Tidjani A, Lounnas M, Hide M, Benavides J, Somasse C, et al. Fecal carriage of extended-spectrum βlactamase-producing Enterobacteriaceae in hospital and community settings in Chad. Antimicrob Resist Infect Control. 2019;8:169.
Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M βlactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017;72(8):2145–55.
Mendonca N, Leitao J, Manageiro V, Ferreira E, Canica M. Spread of extended-spectrum b-lactamase CTX-M-producing Escherichia coli clinical isolates in community and nosocomial environments in Portugal. Antimicrob Agents Chemother. 2007;51:1946–55.
Hanson ND, Moland ES, Hong SG, Propst K, Novak DJ, Cavalieri SJ. Surveillance of community-based reservoirs reveals the presence of CTX-M, imported AmpC, and OXA-30 b-lactamases in urine isolates of Klebsiella pneumoniae and Escherichia coli in a U.S. community. Antimicrob Agents Chemother. 2008;52:3814–16.
Babu R, Kumar A, Karim S, Warrier S, Nair SG, Singh SK, et al. Faecal carriage rate of extended-spectrum β-lactamase-producing Enterobacteriaceae in hospitalised patients and healthy asymptomatic individuals coming for health check-up. J Glob Antimicrob Resist. 2016;6:150–3.
Livermore DM, Hawkey PM. CTX-M: changing the face of ESBLs in the UK. J Antimicrob Chemother. 2005;56:451–4.
Moumouni A, Diagbouga S, Nadembèga C, Metuor Dabire A, Ouattara AK, Zohoncon T, et al. Quinolone Resistance (qnr) genes in fecal carriage of extended Spectrum beta-lactamases producing Enterobacteria isolated from children in Niger. Curr Res Microbiol Biotechnol. 2017;5(1):953–7.
Cattoir V, Weill F-X, Poirel L, Fabre L, Soussy C-J, Nordmann P. Prevalence of qnr genes in Salmonella in France. J Antimicrob Chemother. 2007;59:751–4.
Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005;56:463–9.
Acknowledgements
The authors sincerely thank the “Réseau de Recherche sur les Maladies Entériques à Potentiel Épidémique en Afrique de l’Ouest (REMENTA)” and the “Centre National de Recherche et de Formation sur le Paludisme (CNRFP)/Ouagadougou, Burkina Faso” for technical assistance.
Funding
No funding was received for this study.
Author information
Authors and Affiliations
Contributions
R.D was responsible for initiation of the study and data analysis. Laboratory investigations were performed by RD and AK1 under the guidance of IS, AST, AGS and NB. OT, AK2, WADK, AST, NKG, AAK, AGS and NB participated in data analysis and preparation of the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Permission to conduct the study was obtained from the hospital authorities of Burkina Faso, and informed verbal consent was obtained from the parents/guardians of every child before sample collection. The National Ethical Committee (s) of Burkina Faso (N ° 2009-39) approved the study protocol.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Dembélé, R., Konaté, A., Traoré, O. et al. Extended spectrum beta-lactamase and fluoroquinolone resistance genes among Escherichia coli and Salmonella isolates from children with diarrhea, Burkina Faso. BMC Pediatr 20, 459 (2020). https://doi.org/10.1186/s12887-020-02342-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12887-020-02342-z






 = Salmonella.
= Salmonella.  = Escherichia coli
= Escherichia coli