Abstract
Background
Different moderrn methodologies are presently available to analyze meiotic chromosomes. These methods permit investigation of the behavior of chromosomes in the normal complement and of sex and B chromosomes, two special types of chromosomes that are associated with the A complement and are present in many organisms, including fishes. However, meiotic studies are still scarce in fishes, considering the wide number of species in this group.. Here, we describe a new protocol for the visualization of the synaptonemal complex in spermatocytes and oocytes of fishes and to the sequential use of the technique with other procedures and techniques such as immunodetection of the synaptonemal complex protein with a specific antibody and co-detection of DNA sequences by FISH.
Results
The meiotic surface-spreading protocol used in the present proposal worked well in representative species of four fish orders and was useful in obtaining good results even in small specimens. Fish-specific antibodies and commercial products worked similarly well to detect synaptonemal complex (SC) proteins. The sequential application of fluorescence in situ hybridization using specific probes showed clear signals associated with the SC structures identified by immunostaining.
Conclusion
Here, we provide a useful and applicable immunofluorescent protocol for the visualization of synaptonemal complex proteins in the meiotic cells of fishes in surface-spreading preparations. Furthermore, this technique allows for the sequential application of other cytogenetic procedures.
Similar content being viewed by others
Background
During meiosis, specific events are required to promote genetic diversity and ensure the segregation of homologous chromosomes. These events, including synapsis and meiotic recombination between homologous chromosomes, cause sequential structural changes in chromosomes [1]. Thus, studies on these structural elements can provide important information about pairing and synapsis processes involving homologous chromosomes [2], as well as about the evolution of protein components of the synaptonemal complex (SC) among vertebrates [3].
To accomplish reductional segregation, homologous chromosomes must recognize one another and identify their partners, forming a meiosis-specific organizational structure known as the SC [4,5], which is involved in the chromosome pairing process.
The SC consists of three basic structural filamentous proteins types: two axial or lateral elements connected to the chromatin fibers of chromosomes, one central layer element, and transverse filaments that connect the central layer and the lateral elements [6]. In mammals, seven proteins are described as part of the SC: SYCP2 and SYCP3, forming the lateral elements [7,8]; SYCP1, which is the unique protein that joins the transversal elements [9] and four small proteins, SYCE1, SYCE2, SYCE3 and Tex12, which are specific to the central element [10–12].
In fishes, the presence of a variety of different chromosome polymorphism types, including sex chromosomes and supernumerary chromosomes, has sparked interest in finding information that could lead to a better understanding of the evolutionary processes related to the origin and structure of these chromosomes. However, studies that assess the structure or behavior of chromosomes during meiosis are restricted to a few species, including Danio rerio [13], Acipenser transmontanus [14], Oryzias latipes [15] and Oncorhynchus mykiss [16]. In some works, meiotic chromosomes were analyzed either to compare the pairing processes of chromosomes between closely related species [17,18] or to identify and study the evolution, behavior and origin of sex chromosomes [16,17,19] and B chromosomes [20,21]. The methodology applied in these studies can be difficult to transfer and use in certain groups of fishes in which the pachytene stage occurs in the early phase of the development [4]; in such fishes, the specimens are too small to provide reasonable amounts of tissue to be studied. Another limitation in the application of the technique is the necessity of using electron microscopy to visualize the SC, which is a laborious task and frequently makes it difficult to obtain viable and clear results. Finally, the techniques are not appropriate or not designed to be sequentially used with other cytogenetic procedures.
Here, we describe a new and efficient protocol for the visualization of the SC in spermatocytes and oocytes of fishes adapted from the protocols described by Van Eenennaam et al. [14] and Campos-Ramos et al. [19]. We also demonstrate that the new methodproposed here is effective for species of several groups and even for use in combination with other methods, such as immunodetection of the SYCP3 protein with a specific antibody and co-detection of DNA sequences by fluorescence in situ hybridization (FISH).
Results
Meiotic preparations from different species of fishes such as Oreochromis niloticus, Eigenmannia sp2, Astyanax paranae, and Characidium gomesi were obtained with application of the surface-spreading protocol proposed in this study (Figure 1), highlighting the replicability of the method. Additionally, the results of SYCP3 SC protein immunodetection using a fish-specific antibody were similar to those obtained using mammalian antibodies (Figure 2).
Synaptonemal complex detection with silver stain and with anti-medaka SYCP3 antibody. Meiotic nuclear microspreads in oocytes of Astyanax paranae (a, b, c, d). Spermatocytes of Eigenmannia sp2 (e, f, g, h), Characidium gomesi (i, j, k, l) and Oreochromis niloticus (m, n, o, p). spermatocytes and oocytes were conterstained by DAPI. Bar = 10 μm.
Sequential detection of specific DNA sequences encoding histone H1 by FISH on surface-spreading preparations for SC identification showed clear signals associated with the SC complexes (Figure 3a,b,c), on sites corresponding to specific bivalents. Similarly, chromosome painting with a B chromosome probe revealed the presence of fluorescent signals associated with the SC complex corresponding to this chromosome (Figure 3d,e,f).
FISH hybridization after immunodetection of the synaptonemal complex with anti-medaka SYCP3 antibody. Chromosome painting with specific probes for B chromosome in Characidium gomesi (a, b, c) and histone H1 probe hybridization in Astyanax paranae (d, e, f). In c and f, FISH hybridization signs are visualized on the specific bivalent chromosomes. Bar = 10 μm.
Discussion
The versatility and repeatability of this method coupled with the applicability of this method with complementary cytogenetic techniques greatly increased the resolution capability of the technique to study the composition, structure, and behavior of meiotic chromosomes. The protocol described here allows for the use of fish-specific or commercial, mammalian-derived antibodies to detect protein components of the SC in fishes following pretreatment with citrate buffer, which exposes the protein complexes and allows for antibody binding. The ability to use either fish-specific or mammalian-derived antibodies highlights the state of meiotic protein conservation among vertebrates [15,20]. The preparations obtained using this protocol can also be sequentially used for other techniques such as silver nitrate staining, immunostaining, and fluorescence in situ hybridization to identify the localization of DNA markers. The use of FISH combined with immunostaining in these preparations allows for simultaneous visualization of the pairing behavior of bivalent chromosomes, unpaired regions, and the location of molecular markers or the painting of whole chromosomes during the pachytene stage [21]. Remarkably, this technique is also very useful for small specimens because it can be adapted for the specific volume of the cell suspension for each specimen; further, the number of slides per volume of cell suspension can be altered.
To study meiosis in organisms in the pachytene stage, knowledge of the exact sexual maturation phase of the organisms is essentially required for obtaining good SC preparations in males and females. The pachytene stage occurs in the initial reproductive steps of some species and can be visualized in the early stages of gonadal development [22,23]. Some species are generally too small in this stage, leading to difficulty obtaining a reasonable amount of tissue [24–26]. In fishes, some species breed throughout the year, while others breed only once per year or at certain times during the year, and environmental changes can directly influence the period of gonadal maturation [27–30]. Thus, the main challenge this method serves to successfully address is to provide a perfect understanding of the timing of the meiotic cycle of these species.
Conclusions
The results reported here demonstrate the repeatability and versatility of the protocol for immunodetection of SC proteins in meiotic surface-spreading preparations of fishes. The possibility of using this SC immunodetection technique with the sequential application of other cytogenetic methodologies, including FISH using DNA probes for specific sequences or entire chromosomes, can enhance the resolution of fishes meiotic studies and make possible the identification of chromosomal segments or entire chromosomes. Thus, this method improves our ability to analyze the meiotic behavior of sex chromosomes, B chromosomes and specific chromosome sites during the early stages of meiosis.
Methods
Animal Sampling
This protocol was tested in five fishes species representing the orders Perciformes (Oreochromis niloticus), Gymnotiformes (Eigenmannia sp1, Eigenmannia sp2) and Characiformes (Astyanax paranae and Characidium gomesi).
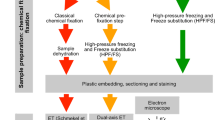
Surface-spreading protocol
Protocol
-
a.
Sacrifice individuals with an overdose of benzocaine.
-
b.
Remove the gonads (testes or ovaries) and immerse them in 200 μl cold Hank’s solution (Sigma) in a Petri dish. The volume of saline solution should represent threefold the tissue volume.
-
c.
Mince tissue to obtain a fine cell suspension.
-
d.
Transfer cell suspension to a 1.5 ml tube and decant for 30 min.
-
e.
Place a drop of cell suspension (approximately 20 μl) in the middle of a clean, dry slide, avoiding the presence of cell debris.
-
f.
Add 2 drops of 0.2 M sucrose and 2 drops of 0.2% Triton X-100 onto the cells; all the solutions must be adjusted to pH 8.5 with 0.1 M sodium tetraborate.
-
g.
Incubate the preparations containing the testis cell or ovarian cell suspensions for 4 min or 7 min, respectively.
-
h.
Fix the cells on the slides by adding 10 drops of 4% paraformaldehyde solution (pH 8.5, adjusted with 0.1 M sodium tetraborate) for 15 min.
-
i.
Remove excess liquid with absorbent paper and let dry at room temperature for 3 hr.
-
j.
Wash 3 times with 0.08% Photo Flo 200 (Kodak) solution and dry in a vertical position.
-
k.
Assess the quality of the preparations by silver nitrate staining according to the protocol described by Howell & Black [31].
-
l.
Store preparations at -20°C in the deep freezer.
Immunostaining
-
a.
Wash the slides three times in 1× PBS for 5 min; do not allow the slides to dry.
-
b.
Antigen recovery*: immerse the slides in 0.01 M citrate buffer (pH 6), preheated to 95°C for 20 min.
-
c.
Allow cooling at room temperature for 20 min.
-
d.
Wash three times in 1× PBS for 2 min each.
-
e.
Incubate with primary antibody (1/100 in 1× PBS), add 25 μl per slide, and cover with a coverslip for 1.5 hr at 37°C in a humid chamber*.
-
f.
Wash three times in 1× PBS for 5 min each (room temperature).
-
g.
Incubate with secondary antibody (1/100 in 1× PBS) by adding 25 μl per slide with a coverslip for 40 min at 37°C.
-
h.
Wash three times 1× PBS, for 5 min each.
-
i.
Mount preparations under coverslips with 15 μL. Vectashield/DAPI.
* The time and temperature for primary antibody incubation can be modified, using either 3 hr. at room temperature or overnight at 4°C.
Sequential FISH and chromosome painting
Following immunodetection, preparations were submitted to FISH. However, as FISH can dim the immunodetection signal, images were taken before this next step was conducted, to be combined with the final image after FISH. The final image composition was performed using image editor software with the double-FISH images, using a uniform image scale and size. However, the SC detection can be improved during FISH by adding secondary antibody to the hybridization solution.
Probes for histone H1 gene and chromosome painting for B chromosomes were produced and labeled according to Hashimoto et al. [32] and de Silva et al. [33] respectively. FISH was performed under highly stringent conditions using the method described by Pinkel et al. [34], without pretreatment steps. Initially, the slides were washed twince in 2× SSC to remove DAPI (4’, 6-diamidino-2-phenylindole, Vector Laboratories). The chromosomal DNA was denatured in 70% formamide/2× SSC for 5 min at 70°C. For each slide, 30 μl of hybridization solution containing 200 ng of each labeled probe, 50% formamide, 2× SSC and 10% dextran sulfate was denatured for 10 min at 95°C, dropped on the slides and hybridized overnight at 37°C in a 2× SSC moist chamber. Post hybridization, the slides were washed two times in 0.2× SSC/15% formamide for 10 min at 42°C, followed by a second wash three times in 0.1× SSC for 5 min at 60°C and a final wash at room temperature in 4× SSC/0.5% Tween for 5 min. Probe detection was carried out with Avidin-FITC (Sigma) or anti-digoxigenin-rhodamine (Roche) and chromosomes were counterstained with DAPI.
Endnotes
Antigen recovery*: Following pretreatment with citrate buffer, the binding sites for the complex of proteins became most exposed for antibody binding.
References
Tsutsumi M, Kogo H, Kowa-Sugiyama H, Inagaki H, Ohye T, Kurahashi H. Characterization of a novel mouse gene encoding an SYCP3-like protein that relocalizes from the XY body to the nucleolus during prophase of male meiosis I. Biol Reprod. 2011;85:165–71.
Takeo S, Lake CM, Morais-de-Sá E, Sunkel CE, Hawley RS, Hawley RS. Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr Biol. 2011;21:1845–51.
Bisig C, Guiraldelli M, Kouznetsova A, Scherthan H, Höög C, Dawson D, et al. Synaptonemal complex components persist at centromeres and are required for homologous centromere pairing in mouse spermatocytes. Plos Genetics. 2012;8:1–13.
Crichton J, Playfoot C, Adams I. The Role of Chromatin modifications in progression through mouse meiotic prophase. J Genet Genomics. 2014;41:97–106.
Zhang J, Zhang BH, Su H, Su O, Birchler J, Han F. Molecular mechanisms of homologous chromosome pairing and segregation in plants. J Genet Genomics. 2014;41:117–23.
Heyting C. Synaptonemal complexes: structure and function. Curr Opin Cell Biol. 1996;8:389–96.
Lammers J, Offenberg H, Aalderen M, Vink A, Dietrich A, Heyting C. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol Cell Biol. 1994;14:1137–46.
Offenberg H, Schalk JA, Meuwissen RL, van Aalderen M, Kester HA, Dietrich AJ, et al. a major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res. 1998;26:2572–9.
Meuwissen R, Offenberg H, Dietrich A, Riesewijk A, Iersel M, Heyting C. A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 1992;11:5091.
Costa Y. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J Cell Sci. 2005;118:2755–62.
Hamer G, Gell K, Kouznetsova A, Novak I, Benavente R, Höög C. Characterization of a novel meiosis-specific protein within the central element of the synaptonemal complex. J Cell Sci. 2006;119:4025–32.
Schramm S, Fraune J, Naumann R, Hernandez-Hernandez A, Höög C, Cooke H, et al. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet. 2011;7:e1002088.
Wallace B, Wallace H. Synaptonemal complex karyotype of zebrafish. Heredity. 2003;40:136–40.
Eenennaam A, Murray J, Medrano J. Synaptonemal complex analysis in spermatocytes of white sturgeon, Acipenser transmontanus Richardson (Pisces, Acipenseridae), a fish with a very high chromosome number. Genome. 1998;41:51–60.
Iwai T, Yoshii A, Yokota T, Sakai C, Hori H, Kanamori A, et al. Structural components of the synaptonemal complex, SYCP1 and SYCP3, in the medaka fish Oryzias latipes. Exp Cell Res. 2006;312:2528–37.
Oliveira C, Foresti F, Rigolino M, Tabata Y. Synaptonemal complex analysis in spermatocytes and oocytes of rainbow trout, Oncorhynchus mykiss (Pisces, Salmonidae): the process of autosome and sex chromosome synapsis. Chromosome Res. 1995;3:182–90.
Campos-Ramos R, Harvey S, McAndrew B, Penman D. An investigation of sex determination in the Mozambique tilapia, Oreochromis mossambicus, using synaptonemal complex analysis, FISH, sex reversal and gynogenesis. Aquaculture. 2003;221:125–40.
Iwai T, Sakai C, Konno F, Yamashita M. Interspecific Medaka Hybrids as Experimental Models for Investigating Cell Division and Germ Cell Development, Chapter 19. In: Medaka: Model for Organogenesis, Human Diseases and Evolution. Springer; 2011. p. 287–303.
Campos-Ramos R, Harvey S, Masab J. Identification of putative sex chromosomes in the blue tilapia, Oreochromis aureus, through synaptonemal complex and FISH analysis. Genetica. 2001;111:140–53.
Baier A, Alsheimer M, Benavente R. Synaptonemal complex protein SYCP3: Conserved polymerization properties among vertebrates. Biochim Biophys Acta. 2007;1774:595–602.
Ocalewicz K, Mota-Velasco J, Campos-Ramos R, Penman D. FISH and DAPI staining of the synaptonemal complex of the Nile tilapia (Oreochromis niloticus) allow orientation of the unpaired region of bivalent 1 observed during early pachytene. Chromosome Res. 2009;17:773–82.
Quagio-Grassiotto I, Grier H, Mazzoni T, Nóbrega R, Amorim J. Activity of the ovarian germinal epithelium in the freshwater catfish, Pimelodus maculatus (Teleostei: Ostariophysi: Siluriformes): germline cysts, follicle formation and oocyte development. J Morphol. 2011;272:1290–306.
Schulz RW, França LR, Lareyre J, LeGac F, Chiarini-Garcia H, Nobrega RH, et al. Spermatogenesis in fish. Gen Comp Endocrinol. 2010;165:390–411.
Schreck CB. Stress and fish reproduction: the roles of allostasis and hormesis. Gen Comp Endocrinol. 2010;165:549–56.
Batlouni S, Romagosa E, Borella M. The reproductive cycle of male catfish Pseudoplatystoma fasciatum (Teleostei, Pimelodidae) revealed by changes of the germinal epithelium. Anim Reprod Sci. 2006;96:116–32.
Patiño R, Sullivan C. Ovarian follicle growth, maturation, and ovulation in teleost fish. Fish Physiol Biochem. 2002;26:57–70.
Kirschbaum F. Reproduction of the weakly electric fish Eigenmannia virescens (Rhamphichtyidae, Teleostei) in captivity: I. Control of gonadal recrudescence and regression by environmental factors. Behav Ecol Sociobiol. 2008;4:331–55.
Giora J, Fialho CB. Reproductive biology of weakly electric fish Eigenmannia trilineata López and Castello, 1966 (Teleostei, Sternopygidae). Brazilian Arch Biol Technol. 2009;52:617. doi:10.1590/S1516-89132009000300014.
de Carvalho PA, Paschoalini AL, Santos GB, Rizzo E, Bazzoli N. Reproductive biology of Astyanax fasciatus (Pisces: Characiformes) in a reservoir in southeastern Brazil. J Appl Ichthyol. 2009;25:306–13.
Vieira A, Melo R, Santos G, Bazzoli N. Reproductive biology of the peacock bass Cichla piquiti (Perciformes: Cichlidae), an exotic species in a Neotropical reservoir. Neotropical Ichthyology. 2009;7:745–50.
Howell W, Black D. Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia. 1980;36:1014–5.
Hashimoto D, Ferguson-Smith M, Rens W, Prado F, Foresti F, Porto-Foresti F. Cytogenetic mapping of H1 histone and ribosomal RNA genes in hybrids between catfish species Pseudoplatystoma corruscans and Pseudoplatystoma reticulatum. Cytogenet Genome Res. 2013;139(2):102–6.
de Silva DMZA, Pansonato-Alves JC, Utsunomia R, Araya-Jaime C, Ruiz-Ruano FJ, Daniel SN, et al. Delimiting the origin of a B chromosome by FISH mapping, chromosome painting and DNA sequence analysis in Astyanax paranae (Teleostei, Characiformes). PloS One. 2014;9(4):e94896. doi: 10.1371/journal.pone.0094896.
Pinkel D, Straume T, Gray J. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci. 1986;83:2934–8.
Acknowledgments
This study was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors of the paper entitled “Surface-spreading technique of meiotic cells and immunodetection of synaptonemal complex proteins in teleostean fishes” (MS ID: 1053723853148885), declare that they have no competing interests of any kind.
Authors’ contributions
CAJ conceived, designed, and carried out the experiments, analyzed the data, and drafted the manuscript. EAS carried out the experiments, analyzed the data and drafted the manuscript. DMZAS carried out the experiments, analyzed the data and drafted the manuscript. MY and TI produced the fish-specific SYCP3 antibody and drafted the manuscript. CO and FF conceived of the study, participated in its design and coordination and helped to draft the manuscript. All of the authors read and approved the final manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Araya-Jaime, C., Serrano, É.A., de Andrade Silva, D.M.Z. et al. Surface-spreading technique of meiotic cells and immunodetection of synaptonemal complex proteins in teleostean fishes. Mol Cytogenet 8, 4 (2015). https://doi.org/10.1186/s13039-015-0108-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-015-0108-9