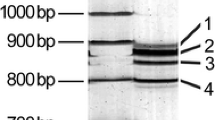
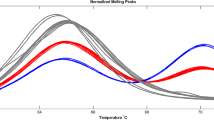
Abstract
Allele mining is a method used to find undiscovered natural variations or induced mutations in a plant, and has become increasingly important as more genomic information is available in plants. A high-throughput method is required to facilitate the identification of novel alleles in a large number of samples. In this paper we describe the application of a high-resolution melting (HRM) method to detect natural variations and ethyl methane sulfonate (EMS)-induced mutations in Capsicum. We have scanned single polymorphic mutations in the first exon of the eIF4E gene, wherein the mutations confer resistance to potyviruses. Sixteen allelic variations out of 248 germplasm collections were identified using HRM analysis, and one accession carrying an allelic variation (pvrHRM1 13) was confirmed to be resistant to the TEV-HAT strain. In addition, five single polymorphic mutations in the eIF4E gene were identified in an EMS-induced mutant population. These results demonstrate that HRM allows for the rapid identification of new allelic variants in both natural and artificial mutant populations.





Similar content being viewed by others
References
Bhullar NK, Street K, Mackay M, Yahiaoui N, Keller B (2009) Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc Natl Acad Sci USA 106:9519–9524
Caldwell DG, McCallum N, Shaw P, Muehlbauer GJ, Marshall DF, Waugh R (2004) A structured mutant population for forward and reverse genetics in Barley (Hordeum vulgare L.). Plant J 40:143–150
Chagne D, Gasic K, Crowhurst RN, Han Y, Bassett HC, Bowatte DR, Lawrence TJ, Rikkerink EH, Gardiner SE, Korban SS (2008) Development of a set of SNP markers present in expressed genes of the apple. Genomics 92:353–358
Charron C, Nicolai M, Gallois JL, Robaglia C, Moury B, Palloix A, Caranta C (2008) Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J 54:56–68
Croxford AE, Rogers T, Caligari PD, Wilkinson MJ (2008) High-resolution melt analysis to identify and map sequence-tagged site anchor points onto linkage maps: a white lupin (Lupinus albus) map as an exemplar. New Phytol 180:594–607
Dong C, Vincent K, Sharp P (2009) Simultaneous mutation detection of three homoeologous genes in wheat by high resolution melting analysis and mutation surveyor. BMC Plant Biol 9:143–154
Gady AL, Hermans FW, Van de Wal MH, van Loo EN, Visser RG, Bachem CW (2009) Implementation of two high through-put techniques in a novel application: detecting point mutations in large EMS mutated plant populations. Plant Meth 5:13–26
Gao Z, Eyers S, Thomas C, Ellis N, Maule A (2004) Identification of markers tightly linked to sbm recessive genes for resistance to Pea seed-borne mosaic virus. Theor Appl Genet 109:488–494
Glaszmann JC, Kilian B, Upadhyaya HD, Varshney RK (2010) Accessing genetic diversity for crop improvement. Curr Opin Plant Biol 13:167–173
Greene EA, Codomo CA, Taylor NE, Henikoff JG, Till BJ, Reynolds SH, Enns LC, Burtner C, Johnson JE, Odden AR (2003) Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164:731–740
Hofinger BJ, Jing HC, Hammond-Kosack KE, Kanyuka K (2009) High-resolution melting analysis of cDNA-derived PCR amplicons for rapid and cost-effective identification of novel alleles in barley. Theor Appl Genet 119:851–865
Hwang J, Li J, Liu WY, An SJ, Cho H, Her NH, Yeam I, Kim D, Kang BC (2009) Double mutations in eIF4E and eIFiso4E confer recessive resistance to Chilli veinal mottle virus in pepper. Mol Cell 27:329–336
Jeong HJ, Jo YD, Park SW, Kang BC (2010) Identification of Capsicum species using SNP markers based on high resolution melting analysis. Genome 53:1029–1040
Johnson RC (2008) Gene banks pay big dividends to agriculture, the environment, and human welfare. PLoS Biol 6:e148
Kang BC, Yeam I, Frantz JD, Murphy JF, Jahn MM (2005) The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J 42:392–405
Kumar GR, Sakthivel K, Sundaram RM, Neeraja CN, Balachandran SM, Rani NS, Viraktamath BC, Madhav MS (2010) Allele mining in crops: prospects and potentials. Biotechnol Adv 28:451–461
Lehmensiek A, Sutherland MW, McNamara RB (2008) The use of high resolution melting (HRM) to map single nucleotide polymorphism markers linked to a covered smut resistance gene in barley. Theor Appl Genet 117:721–728
Mackay JF, Wright CD, Bonfiglioli RG (2008) A new approach to varietal identification in plants by microsatellite high resolution melting analysis: application to the verification of grapevine and olive cultivars. Plant Meth 4:8–17
Menda N, Semel Y, Peled D, Eshed Y, Zamir D (2004) In silico screening of a saturated mutation library of tomato. Plant J 38:861–872
Minoia S, Petrozza A, D’Onofrio O, Piron F, Mosca G, Sozio G, Cellini F, Bendahmane A, Carriero F (2010) A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res Notes 3:69–77
Monzingo AF, Dhaliwal S, Dutt-Chaudhuri A, Lyon A, Sadow JH, Hoffman DW, Robertus JD, Browning KS (2007) The structure of eukaryotic translation initiation factor-4E from wheat reveals a novel disulfide bond. Plant Physiol 143:1504–1518
Muleo R, Colao MC, Miano D, Cirilli M, Intrieri MC, Baldoni L, Rugini E (2009) Mutation scanning and genotyping by high-resolution DNA melting analysis in olive germplasm. Genome 52:252–260
Park SW, An SJ, Yang HB, Kwon JK, Kang BC (2009) Optimization of high resolution melting analysis and discovery of single nucleotide polymorphism in Capsicum. Hort Environ Biotechnol 50:31–39
Reed GH, Wittwer CT (2004) Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem 50:1748–1754
Rubio M, Nicolai M, Caranta C, Palloix A (2009) Allele mining in the pepper gene pool provided new complementation effects between pvr2-eIF4E and pvr6-eIF(iso)4E alleles for resistance to pepper veinal mottle virus. J Gen Virol 90:2808–2814
Stephenson P, Baker D, Girin T, Perez A, Amoah S, King GJ, Ostergaard L (2010) A rich TILLING resource for studying gene function in Brassica rapa. BMC Plant Biol 10:62
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Till BJ, Reynolds SH, Weil C, Springer N, Burtner C, Young K, Bowers E, Codomo CA, Enns LC, Odden AR (2004) Discovery of induced point mutations in maize genes by TILLING. BMC Plant Biol 4:12
Triques K, Piednoir E, Dalmais M, Schmidt J, Le Signor C, Sharkey M, Caboche M, Sturbois B, Bendahmane A (2008) Mutation detection using ENDO1: application to disease diagnostics in humans and TILLING and Eco-TILLING in plants. BMC Mol Biol 9:42
Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H (2008a) Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet 40:1370–1374
Wang M, Allefs S, van den Berg RG, Vleeshouwers VG, van der Vossen EA, Vosman B (2008b) Allele mining in Solanum: conserved homologues of Rpi-blb1 are identified in Solanum stoloniferum. Theor Appl Genet 116:933–943
Wang N, Wang Y, Tian F, King GJ, Zhang C, Long Y, Shi L, Meng J (2008c) A functional genomics resource for Brassica napus: development of an EMS mutagenized population and discovery of FAE1 point mutations by TILLING. New Phytol 180:751–765
Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ (2003) High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem 49:853–860
Wu JL, Wu C, Lei C, Baraoidan M, Bordeos A, Madamba MR, Ramos-Pamplona M, Mauleon R, Portugal A, Ulat VJ (2005) Chemical- and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol Biol 59:85–97
Wu SB, Wirthensohn MG, Hunt P, Gibson JP, Sedgley M (2008) High resolution melting analysis of almond SNPs derived from ESTs. Theor Appl Genet 118:1–14
Wu SB, Tavassolian I, Rabiei G, Hunt P, Wirthensohn MG, Gibson JP, Ford CM, Sedgley M (2009) Mapping SNP-anchored genes using high-resolution melting analysis in almond. Mol Genet Genomics 282:273–281
Yeam I, Kang BC, Lindeman W, Frantz JD, Faber N, Jahn MM (2005) Allele-specific CAPS markers based on point mutations in resistance alleles at the pvr1 locus encoding eIF4E in Capsicum. Theor Appl Genet 112:178–186
Yeam I, Cavatorta JR, Ripoll DR, Kang BC, Jahn MM (2007) Functional dissection of naturally occurring amino acid substitutions in eIF4E that confers recessive potyvirus resistance in plants. Plant Cell 19:2913–2928
Yu JB, Bai GH, Cai SB, Dong YH, Ban T (2008) New Fusarium head blight-resistant sources from Asian wheat germplasm. Crop Sci 48:1090–1097
Acknowledgments
This work was carried out by a grant of the Technology Development Program and the Agricultural Research Center Program of Agriculture and Forestry, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea. This research was supported by Technology Development Program for Agriculture and Forestry, Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jeong, HJ., Kwon, JK., Pandeya, D. et al. A survey of natural and ethyl methane sulfonate-induced variations of eIF4E using high-resolution melting analysis in Capsicum . Mol Breeding 29, 349–360 (2012). https://doi.org/10.1007/s11032-011-9550-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-011-9550-5