Abstract
Jasmonates control many aspects of plant biological processes. They are important for regulating plant responses to various biotic and abiotic stresses, including drought, which is one of the most serious threats to sustainable agricultural production. However, little is known regarding how jasmonate ZIM-domain (JAZ) proteins mediate jasmonic acid signals to improve stress tolerance in cotton. This represents the first comprehensive comparative study of TIFY transcription factors in both diploid A, D and tetraploid AD cotton species. In this study, we identified 21 TIFY family members in the genome of Gossypium arboretum, 28 members from Gossypium raimondii and 50 TIFY genes in Gossypium hirsutum. The phylogenetic analyses indicated the TIFY gene family could be divided into the following four subfamilies: TIFY, PPD, ZML, and JAZ subfamilies. The cotton TIFY genes have expanded through tandem duplications and segmental duplications compared with other plant species. Gene expression profile revealed temporal and tissue specificities for TIFY genes under simulated drought conditions in Gossypium arboretum. The JAZ subfamily members were the most highly expressed genes, suggesting that they have a vital role in responses to drought stress. Over-expression of GaJAZ5 gene decreased water loss, stomatal openings, and the accumulation of H2O2 in Arabidopsis thaliana. Additionally, the results of drought tolerance assays suggested that this subfamily might be involved in increasing drought tolerance. Our study provides new data regarding the genome-wide analysis of TIFY gene families and their important roles in drought tolerance in cotton species. These data may form the basis of future studies regarding the relationship between drought and jasmonic acid.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cotton is the most important fiber and oil crop worldwide, and its production is threatened by abiotic stresses, such as drought and salinity stresses (Grayson 2013). The International Service for the Acquisition of Agri-biotech Application reported that in Australia, the area on which cotton was grown decreased by 200,000 hectares between 2013 and 2014 because of an ongoing drought (http://www.isaaa.org/). Therefore, breeding for drought-resistant cotton should be a high priority for plant biotechnology programs. Phytohormones, such as auxin, gibberellin, and abscisic acid, considerably affect plant growth, development, aging, dormancy, and stress resistance. Jasmonic acid (JA) is considered as a new plant hormone, and there is interest regarding whether it can be used to increase stress tolerance in cotton (Kumar et al. 2014).
Jasmonic acid is a small oxylipin signaling molecule that is ubiquitous in the plant kingdom. It regulates plant responses to various biotic and abiotic stresses, such as wounding, UV light, water deficit, pathogens, and ozone. It is also involved in a wide range of plant developmental processes, including pollen and stamen development, vegetative growth control, anthocyanin accumulation, and senescence (Conconi et al. 1996; McConn and Browse 1996; Creelman and Mullet 1997; Rao et al. 2000; Berger 2002; Schommer et al. 2008; Cheng et al. 2009; Ren et al. 2009; Qi et al. 2011; Wang et al. 2013).
COI1 is the F-box protein subunit of SCFCOI1 (ubiquitin ligase complex), which is a member of the Skip/Cullin/F-box family of E3 ubiquitin ligases that functions as a central conserved component of the JA signaling pathway in Arabidopsis thaliana and tomato (Xie et al. 1998; Chini et al. 2007; Thines et al. 2007). The substrate specificity of SCFCOI1 is mainly based on the specificity of the F-box protein. The substrate is ubiquitinated by E3 ubiquitin ligases, and subsequently degraded by the 26S proteasome (Moon et al. 2004).
The jasmonate ZIM-domain (JAZ) proteins were identified as a substrate of SCFCOI1 by Thines et al. (2007). The JAZ proteins are localized to the nucleus, although they do not contain a DNA-binding domain (Grunewald et al. 2009). These proteins repress jasmonate responses by interacting with the basic helix-loop-helix (bHLH) transcription factors MYC2 and MYC3, which bind to DNA sequences and regulate downstream gene expression (Lorenzo et al. 2004; Chini et al. 2007; Grunewald et al. 2009; Cheng et al. 2011). SCFCOI1/JAZ/MYC is a three-component core complex of the JA signaling pathway. Because SCFCOI1 is highly conserved, JA is believed to have diverse functions that depend on JAZ proteins and MYC transcription factors.
In 2007, proteins containing a zinc finger protein expressed in inflorescence meristem (ZIM) domain were discovered in a study of JA responses in Arabidopsis thaliana (Nishii et al. 2000). Because of the presence of a conserved motif (i.e., TIFYXG), the ZIM domain was renamed as the TIFY domain (Vanholme et al. 2007). The JAZ proteins belong to the TIFY family, and have been identified as a key regulator of JA (Chini et al. 2007; Thines et al. 2007). The well-characterized TIFY genes have been studied for their vital role in JA-related mechanisms. Several studies demonstrated that when plants are treated with JA and other stresses, such as pathogens, drought, cold, or salinity, the TIFY genes provide protection by inducing a differential negative feedback loop mechanism that returns plants to their normal state (Chini et al. 2007; Thines et al. 2007; Chung et al. 2008; Demianski et al. 2012).
Previous studies on JAZ proteins in the model plant A. thaliana confirmed their importance as regulators of JA responses. Recent studies have focused on rice and grapevines. Several rice and grape genes that might influence tolerance to various types of abiotic stress, such as drought, salinity, and low temperature, have been suggested (Ye et al. 2009; Zhang et al. 2012). Currently, there is limited information regarding JA expression and the functions of TIFY family genes in cotton.
Cotton is valued for its role in fiber production, and is a vital commodity in the global economy. The availability of a sequenced Gossypium spp. genome enables genome-wide identification and computational analysis of the TIFY gene family. In this study, we report the analysis of the TIFY genes from Gossypium arboretum, Gossypium raimondii and Gossypium hirsutum pertaining to genomic organization, gene structures and the phylogenetic tree analysis. Although G. arboreum (A genome) produces inferior fiber, it exhibits drought tolerance, disease resistance, and can grow in desiccated areas (Mehetre et al. 2003). So, the TIFY gene family expression in G. arboretum is observed under drought stress conditions and conduct drought tolerance assays. In addition, the results indicate the over-expression of GaJAZ5 genes enhance drought tolerance in cotton by affecting various stress-related pathways. To the best of our knowledge, this is the first report of a genome-wide analysis of TIFY genes in cotton. The potential roles of TIFY genes in drought stress signaling and JA signaling pathways should be further investigated.
Materials and methods
Identification and motif analysis of the TIFY genes
The DNA and corresponding amino acid sequences of 21 TIFY genes were retrieved from the Genome A Database (http://cgp.genomics.org.cn/), based on the published mRNA-seq data (Zhang et al. 2013). A local BLASTP search was completed to identify complete TIFY genes, using A. thaliana TIFY sequences as queries. A BLASTP search (e value: 1e−10) was used to obtain a dataset of TIFY proteins, including 28 Gossypium raimondii, 50 Gossypium hirsutum, 16 Theobroma cacao sequences, 18 A. thaliana sequences, 14 Ricinus communis sequences, 25 Populus trichocarpa sequences, and 34 Glycine max sequences downloaded from the GenBank (http://ncbi.nlm.nih.gov/genbank) and Phytozome (http://phytozome.jgj.doe.gov/pz/portal.html) databases. To identify all members of the TIFY gene family, previous TIFY gene sequences were analyzed using the Simple Modular Architecture Research Tool (SMART; http://smart.embl-heidelberg.de/) (Letunic et al. 2006) and Pfam (http://pfam.janelia.org/) (Bateman et al. 2004).
Sequence alignment and phylogenetic analysis
Multiple amino acid sequence alignments of TIFY proteins from G. arboreum, G.raimondii, G. hirsutum, T. cacao, A. thaliana, R. communis, P. trichocarpa, and G. max were completed with the ClustalW program (Larkin et al. 2007). Additionally, phylogenetic trees were constructed using MEGA 5.05 software, and evolutionary distances were determined using the p-distance method and 1000 bootstrap samples (Wei et al. 2014), which established the reliability of each tree. Amino acid identities were confirmed using JEMBOSS 1.5. Synonymous (dS) and non-synonymous (dN) substitution rates were calculated with the PAL2 NAL web server (http://www.bork.embl.de/pal2nal/#RunP2N), which uses the CODEMAL program of PAML (Suyama et al. 2006).
Structure analysis and chromosomal localization of the TIFY genes
We used SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1) to identify TIFY domains (Letunic et al. 2012), and the MEME (http://meme.nbcr.net/meme/) program was used to predict motifs. The exon–intron structures of the TIFY genes were determined by comparing the coding sequences and corresponding genomic sequences. The Gene Structure Display Server (http://gsds.cbi.pku.edu.cn) (Hu et al. 2015) was used to analyze the exon–intron structures. Data from the Genome Database and Mapchart software were used to predict the chromosomal locations of the TIFY genes. Adobe Photoshop software was used to visualize the distribution of TIFY genes on the chromosomes.
Gossypium arboreum TIFY gene expression analysis
To determine TIFY gene expression levels and assess their association with drought resistance in leaves, roots, and stems, G. arboreum cv. ‘Shixiya’ seedlings were treated with polyethylene glycol (PEG) to simulate drought conditions and then analyzed by quantitative reverse transcription polymerase chain reaction. After the emergence of three to four leaves, plants were treated with 17 % PEG 6000 or water (control) for 6 h. Roots, stems, and leaves were collected every 30 min for subsequent qRT-PCR analyses. The sample was obtained from at least three individual plants for each stage. An RNAprep Pure Plant kit (Tiangen, China, Beijing) was used to extract total RNA. Reverse transcription reactions were completed using 0.5 μg RNA and an iScript cDNA synthesis kit (Bio-Rad, USA). The ABI Power SYBR Green PCR Master Mix (Applied Biosystems, USA) was used with 5 ng template. First-strand cDNA was synthesized from DNaseI-treated total RNA using 1 μl iScript reverse transcriptase (TaKaRa, Japan), 0.5 μg RNA, and 4 μl 5× Reaction Mix. Samples were incubated at 25 °C for 5 min, followed by 42 °C for 30 min, 85 °C for 5 min, and finally storage at 4 °C. The qRT-PCR was conducted in an optical 384-well plate using an ABI PRISM 7500 real-time PCR system (Applied Biosystems). Gene-specific primers were designed for TIFY gene family members (Table S1). The SYBR Green qRT-PCR reaction consisted of 5 μl 2× SYBR Green PCR buffer, 0.5 μl primers, and 5 ng templates. The final volume was adjusted to 10 μl with double-distilled H2O. The PCR program was as follows: 50 °C for 2 min; 95 °C for 10 min; 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Data were processed using the 2−ΔΔCt method (Livak and Schmittgen 2001). The experiment was repeated three times. A heat map was prepared using Cluster 3.0.
Cloning of the Gossypium arboreum TIFY genes
Total RNA was extracted from G. arboreum seedling tissues using Trizol (Sigma-Aldrich) according to the manufacturer’s instructions. Samples were reverse transcribed using an oligo (dT) primer and SuperScript II reverse transcriptase (Promega, USA). The resulting product was used as a PCR template to amplify the predicted TIFY open reading frames using PrimeSTAR DNA Polymerase. The TIFY cDNA regions were cloned into a T-simple vector (TaKaRa). The accuracy of all clones was confirmed by sequencing. TIFY-specific primers were designed, synthesized, and used to clone the genes. For plant transformations, the TIFY cDNA samples were inserted into the modified pSuper1300 plant transformation vector (provided by Zhen Su from China Agricultural University) under the control of the cauliflower mosaic virus 35S promoter.
Arabidopsis thaliana transformation and screening of transgenic plants
The TIFY-pSuper1300 constructs were introduced into Agrobacterium tumefaciens strain GV3101 cells, which were then used to transform A. thaliana plants according to the floral dip method (Clough and Bent 1998). The resulting T3 generation was used for all subsequent experiments. Total DNA was extracted from G. arboreum tissues with the DNAprep Pure Plant kit (Tiangen, China). Primers were synthesized (forward primer: TTGAATAGATACGCTGACACGC; reverse primer: CTATCCCTTTCTCTTCTCGA) and used along with the following PCR program: 94 °C for 5 min; 32 cycles at 94 °C for 40 s, 60 °C for 40 s, and 72 °C for 50 s.
Effect of polyethylene glycol on TIFY gene expression
The effects of PEG on TIFY expression were evaluated by qRT-PCR. Samples were collected from A. thaliana plants treated with PEG for 4 h. Total RNA was extracted from the samples and reverse transcribed as described to generate cDNA for qRT-PCR analyses.
Drought tolerance assay
Drought tolerance was assessed by transferring 7-day-old plants cultured in Petri dishes to pots (10-cm diameter) filled with a vermiculite:perlite mixture [1:1.5 (v/v)]. Seedlings were cultured for 2 weeks with constant watering before drought conditions were simulated. After 17 days without water, all plants were simultaneously rewatered, and plant re-growth was scored 2 days later. Six plates for individual transgenic lines were used in each repeated experiment, with every plate consisting of 35 g vermiculite:perlite mixture.
Water loss assay
Leaves were harvested from 9-week-old mutant and wild-type (WT) seedlings at the rosette stage. Changes in fresh weight (FW) over time were monitored using an electronic balance, and the rate of water loss was calculated as the loss in fresh weight. Ten plants of each transgenic and WT line were analyzed in this assay, which was replicated three times (Duan et al. 2012). Fully expanded leaves were cut from the plants, and the FW was recorded immediately. Then, the leaves were immersed in distilled water for 0.5 h and the turgid weight was recorded. Finally, the dry weight (DW) was recorded after drying for 6 h at 23 °C in an oven. The water loss rate (%) was calculated as follows: (FW − DW) × 100/FW.
Ratio of open to closed stomata
Leaf samples (without veins or edges) were harvested from 7-day-old mutant and WT seedlings, and treated with 10 % PEG 6000 for 3 h. Samples were observed under a light microscope and the open and closed stomata were counted.
Hydrogen peroxide analysis
Leaves from 10-day-old mutant and WT seedlings were treated with 10 % PEG 6000 for 12 h. The H2O2 content was then determined. Leaves and stems were harvested from 4-week-old mutant and WT seedlings at the rosette stage following treatment with 10 % PEG 6000 or water (control) for 12 h. Samples were treated with the REAL EnVision Detection System (Peroxidase/DAB+, Rabbit/Mouse) (Dako) solution for 8 h at 28 °C, destained with ethanol, and then observed under a stereoscopic microscope.
Results
Genome-wide identification of TIFY genes
Twenty-one TIFY family genes were obtained from the cotton Genome A Database (http://cgp.genomics.org.cn). All of the genes had different Garb numbers resulting from mRNA-seq data (Zhang et al. 2013). Then, we used a Blast program (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/release) based on the multiple sequence alignment results of Arabidopsis TIFY protein sequences to identify TIFY genes in Gossypium arboretum. We used databases containing Gossypium raimondii and Gossypium hirsutum genome sequences of the cotton research institute of Chinese academy of agricultural sciences, anyang, Henan (Li et al. 2015). After removing redundant sequences, we identified 28 TIFY genes in the G. raimondii and 50 TIFY genes in the G. hirsutum with an e value of less than one. We named the genes according to their chromosomal locations and genetic structures (Table 1, Table S2, Table S3 and Fig. S1).
The 21 TIFY family genes had a high similarity index (i.e., 91.94–99.72 %) among G. arboreum (genome A), G. raimondii (genome D), and G. hirsutum (genome AD) genomes (Table 1; Fig. 1). This similarity indicated the genes were highly conserved during the evolution of Gossypium spp., except for unigene 3984 (Fig. S2). These findings indicated that the TIFY genes likely diverged evolutionarily from early terrestrial plants, which involved considerable mutations that resulted in functional divergence. Because of the similarity among the A, D, and AD genomes, and to avoid the complexity of G. hirsutum (tetraploid cotton), we used G. arboreum to study the relationships among TIFY family genes, JA, and drought conditions.
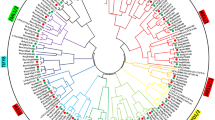
Unrooted neighbor-joining phylogenetic tree of TIFY genes from Gossypium arboreum, Gossypium raimondii, Gossypium hirsutum, Arabidopsis thaliana, Theobroma cacao, Glycine max, Ricinus communis, and Populus trichocarpa. Group I ZML subfamily, Group II TIFY subfamily, Group III PPD subfamily, Group IV JAZ subfamily
Phylogenetic analysis and classification of the TIFY gene family
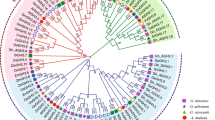
To assess the phylogenetic relationships of the TIFY family among G. arboreum, G. raimondii, and G. hirsutum, A. thaliana, T. cacao, G. max, R. communis, and P. trichocarpa, 206 full-length amino acid sequences were used to construct a phylogenetic tree (Fig. 1). The results indicated that all of the analyzed species diverged from a common ancestor, with cotton diverging the most. The amino acid sequences could be divided into four groups. Using the cotton protein sequences as an example, the protein sequences containing the TIFY and Jas motifs were predicted to belong to the JAZ subfamily. The proteins lacking a conserved PY structure at the C-terminus were predicted to belong to the PPD subfamily. The proteins with TIFY and CCT motifs, and a ZnF_GATA motif, were predicted to belong to the ZML subfamily. The protein contained just the TIFY motif, which suggested it belonged to the TIFY subfamily (Fig. 2a). These classification results were consistent with those based on domain compositions, as described above (Fig. 2b). The structure of the main motif is presented in Fig. 2c. We identified 28 TIFY genes in the G. raimondii genome, which were classified into the TIFY (two genes), PPD (three genes), ZML (eight genes), and JAZ (15 genes) groups (Table S2) (He et al. 2015). Additionally, 50 TIFY genes were identified in the G. hirsutum genome, and were classified into the TIFY (three genes), PPD (five genes), ZML (17 genes), and JAZ (25 genes) groups (Table S3). The G. arboreum TIFY genes were divided into four subfamilies: TIFY (one gene), PPD (two genes), ZML (five genes), and JAZ (13 genes). According to the phylogenetic tree, the JAZ subfamily could be divided into six subgroups and were associated with A. thaliana, T. cacao, G. max, R. communis, and P. trichocarpa (Table 2).
Phylogenetic relationships and gene structures of Gossypium spp. TIFY genes. a Four subfamilies (Group I–IV) are indicated with different colors. Group IV is divided into six subgroups with different colors. A schematic diagram of amino acid motifs is provided on the right side. b Distribution of the conserved motifs in the ZML, PPD, and JAZ subfamilies identified by MEME. Motif locations are indicated at the bottom. c TIFY, Jas, and ZnF_GATA motifs
Analysis of the exon–intron structure of the TIFY genes
To determine the origin of TIFY paralogs, we analyzed the structures of the TIFY genes. Compared with the phylogenetic results, the results of the analyses of exon–intron structures suggested that Groups I–IV had undergone exon deletions. The exon–intron analysis shows that gap of exons in G. arboretum and G. raimondii was smaller, but most the number of exons in tetraploid cotton (G. hirsutum) was the sum of exons in G. arboretum and G. raimondii or similar to G. arboretum and G. raimondii. Take exon–intron analysis in G. arboretum as examples, we made a concrete analysis of exon–intron. Analyses of the exon–intron structure revealed there were 3–12 exons in each of the analyzed TIFY genes in G. arboreum. Eight TIFY genes had six or seven exons, and six TIFY genes had four or five exons. Additionally, GaJAZ6 had the most exons and introns, possibly because of gene duplication events (Fig. 3). Furthermore, 80 % of the ZML subfamily members had seven exons, suggesting that the genomes were highly conserved during evolution. However, the number of exons among JAZ subfamily members ranged from 3 to 12, suggesting that considerable changes had taken place during evolution. Therefore, the proteins in the JAZ subfamily likely have different functions because of a frame shift. This conservation of exon and intron numbers correlated well with the clades identified in the phylogenetic tree for each group. This strongly supports the close evolutionary relationships of TIFY genes in Groups I–IV.
Chromosomal location and evolutionary history of the TIFY gene family
Twenty-one TIFY genes were mapped to 11 G. arboreum chromosomes. There were thirty-eight TIFY genes mapped to 13 chromosomes and the other 12 genes distribution on the 9 scaffolds in G. hirsutum, which were not assemble into chromosomes. All the TIFY genes G. raimondii were located on the scaffolds, which was different with the published articles (He et al. 2015). Take diploid cotton G. arboretum as examples, each chromosome contained at most four genes (Fig. S1). It is believed that the duplication mechanisms (e.g., tandem, segmental, and whole-genome duplications) in eukaryotes are very similar. These mechanisms caused gene families to expand, as observed in G. arboreum (Wang et al. 2012). To infer the possible relationships between TIFY genes and potential gene duplications, we analyzed the occurrence of tandem and large-scale segmental duplications during the evolution of this gene family (Table S4). Among the 21 TIFY genes, six paralogs were identified according to the chromosomal locations. By analyzing the similarities and sequences of the six paralogs, we determined that only one gene had undergone a tandem duplication event (i.e., GaJAZ2/1), implying that this type of duplication was not prevalent during the evolution of the TIFY gene family. Five segmental duplication events (i.e., GaJAZ3/12, GaJAZ9/10, GaZML3/1, GaZML4/2, and GaJAZ11/13) were detected in G. arboreum (Fig. S1). This result suggested that the expansion of the G. arboreum TIFY gene family was mainly due to segmental duplication events.
Nucleotide substitutions in protein-coding genes can be classified as synonymous or non-synonymous. To assess the evolutionary history of the TIFY gene family, d S and d N values were calculated using PAL2NAL. These values can be used to determine whether protein-coding genes have undergone a selection process (de Las et al. 2004). We estimated the d N/d S ratios for the six pairs of duplicated genes (Table S4) and determined they were all less than one. This suggested the duplicated gene pairs experienced a purifying selection process (Goldman and Yang 1994).
Expression analysis of 18 selected Gossypium arboreum TIFY genes
Because TIFY genes have a pivotal role in the JA signaling pathway, we analyzed the expression of 21 G. arboretum TIFY genes in roots, stems, and leaves under drought stress conditions (i.e., 17 % PEG 6000) to determine their functions. Among the 21 genes, 18 (86 %) were expressed in at least one of the three tissues (Fig. 4). Expression of the other three genes was not expressed according to the qRT-PCR analyses.
The expression levels of seven GaJAZ genes (i.e., GaJAZ3, GaJAZ4, GaJAZ5, GaJAZ7, GaJAZ8, GaJAZ9, and GaJAZ13) were highest in leaves at all time points. Treatment with 17 % PEG 6000 resulted in high transcript levels that peaked 1 h after the initiation of drought stress. However, expression levels tended to gradually decrease in the roots. In stems, transcripts of four GaJAZ genes (i.e., GaJAZ2, GaJAZ3, GaJAZ11, and GaJAZ12), GaPPD1, and GaTIFY peaked at 4 h after PEG treatment. Additionally, GaJAZ7 was the most highly expressed gene in all tissues. Overall, the JAZ subfamily members exhibited the highest expression levels in the three analyzed tissues, suggesting GaJAZ genes are important for responses to drought stress.
TIFY gene responses to drought conditions
A schematic representation of a 35S-TIFY gene construct is presented in Fig. 5a. The successful transformation of A. thaliana plants with the TIFY constructs was verified by PCR analyses of marker and target genes. Five independent GaJAZ5 single-copy transgenic lines were generated, and analyzed by genomic PCR to confirm transformation and over-expression (Fig. 5b, c). The GaJAZ5 expression levels varied among the transgenic lines, indicating the gene was successfully introduced into the transformed plants (Fig. 5d).
Plant transformation vector and the expression patterns of the TIFY genes in the transgenic lines. a Schematic representation of the T-DNA region of the pSuper1300-GaJAZ5 binary vector. b PCR analysis of the selected marker gene. WT wild-type, NT non-template, P plasmid, L1, L2, L3, L7, L8 transgenic lines expressing GaJAZ5. c Genomic PCR analysis of the GaJAZ5 gene. d GaJAZ5 expression patterns in transgenic plants. Values are presented as the means of three replicates. Error bars indicate the standard deviations. The histone gene was used as an internal control to normalize gene expression levels
To test whether GaJAZ5 influenced drought resistance, a water loss assay was completed. After withholding water for 17 days, some of the WT plants wilted and died, while only a few GaJAZ5 transgenic plants wilted. After plants were rewatered, the transgenic plants recovered from the drought conditions much better than the WT plants (Fig. 6). These results suggested that GaJAZ5 affects drought tolerance.
The rate of water loss was investigated to further characterize how GaJAZ5 influences water stress tolerance through the maintenance of high relative water content and a reduced rate of water loss. Because of their smaller stomatal apertures (Fig. 7a), the transgenic A. thaliana plants over-expressing GaJAZ5 retained more water than the WT plants following treatment with 10 % PEG 6000. Additionally, the in vitro leaf water loss rate in transgenic lines 3 and 7 was lower than that of WT plants (Fig. 7b).
Under normal conditions, the H2O2 levels in all GaJAZ5-expressing lines were lower than those of the WT plants. Following treatment with 10 % PEG 6000, the H2O2 levels of three lines (especially transgenic line 3) were lower than those of the WT plants (Fig. 7c). This indicated that WT plants accumulated more H2O2 than the GaJAZ5-expressing transgenic plants after treatment with 10 % PEG 6000 (Fig. 7d).
Discussion
Importance of TIFY genes
Survey data indicated drought-affected environments will seriously threaten sustainable agricultural production of crops (http://www.isaaa.org/). Therefore, it is important to identify genes that promote high water use efficiency and that are suitable for plants in desert areas. Cotton is one of the most economically important crops worldwide (Kalivas et al. 2011). Gossypium arboreum (diploid) is drought tolerant, and a better candidate for studying drought tolerance-related genes than G. hirsutum (tetraploid) (Chen et al. 2015). Jasmonic acid regulates plant defense activities by inducing the expression of TIFY genes, which forms part of a negative feedback loop biosynthesis pathway and a signal transduction pathway that controls JAZ protein levels in response to JA and environmental stress (Song et al. 2014). JAZ genes are well-characterized members of the TIFY family with roles in JA-mediated processes, and thus are valuable for stress tolerance research.
Phylogenetic analysis and classification of the TIFY gene family
TIFY homologs are only present in terrestrial plants, and not in green algae or other non-photosynthetic eukaryotes (Vanholme et al. 2007). This suggests that the TIFY family may have originated after aquatic plants evolved to survive on land. Comparative genomic analyses involving different taxa are useful for studying the structure, function, and/or evolution of genomes from uncharacterized taxa (d’Alencon et al. 2010). The TIFY family genes in G. raimondii and G. arboreum are highly conserved, but some G. arboreum genes were lost as the species evolved (Fig. 2). Both G. raimondii and G. arboreum underwent cotton-specific whole-genome duplication events approximately 16.6 million years ago. The modern allotetraploid G. hirsutum species resulted from hybridizations of two (A or D diploid) ancestral species approximately 1.5 million years ago (Li et al. 2015). All TIFY family members in the analyzed species contained four subfamilies, indicating they may have evolved similarly. Thus, we predicted target gene functions and/or evolutions using model plants such as A. thaliana. Previous studies demonstrated that eight AtJAZ genes (i.e., AtJAZ1, AtJAZ2, AtJAZ5, AtJAZ6, AtJAZ7, AtJAZ8, AtJAZ9, and AtJAZ10) were responsive to JA (Thines et al. 2007; Chung et al. 2009). Additionally, other aspects of A. thaliana TIFY gene activities, including alternative splicing (Chung et al. 2009) and interactions with MYC2 (Chini et al. 2007), were systematically analyzed to predict TIFY genes based on A. thaliana syntenic orthologs.
Conservation and divergence: tandem and segmental duplications contributed to the expansion of the TIFY gene family
We detected two sequences with the same Garb number, and another sequence that had two Garb numbers (Zhang et al. 2013). Nevertheless, we identified 21 TIFY genes in the G. arboreum by motif analysis and subsequent molecular cloning, and determined that the cotton TIFY gene family is one of the largest TIFY families reported to date. Preliminary analyses revealed that gene duplication and subsequent divergence events were the main contributors to evolutionary momentum (Ohno et al. 1968; Chothia et al. 2003). Usually, the criteria for inferring a gene duplication event are as follows: the length of the alignment sequence covers ≥80 % of the longest gene, and the similarity of the aligned regions is ≥70 % (Gu et al. 2002; Yang et al. 2008). Two or more identical genes located on the same chromosome result from tandem duplications, whereas gene duplications between different chromosomes are designated as segmental duplications (He et al. 2012). The TIFY genes of A. thaliana and maize have undergone both segmental and tandem duplications, contributing to the expansion of the TIFY gene family. Tandem and segmental duplications of TIFY genes have also occurred in rice (Melotto et al. 2008) and grapevines (Zhang et al. 2012). Our study indicates that the G. arboreum TIFY genes are similar to those of A. thaliana, with tandem and segmental duplication events occurring in both species. Gene duplication followed by functional diversification (i.e., evolutionary changes in expression patterns) has played a vital role in driving the evolutionary processes that increase fitness to new environments (Conrad and Antonarakis 2007; Conant and Wolfe 2008). Future research may affect the topology of the phylogenetic tree in terms of the order of evolutionary distances for different members of the cotton TIFY family. Therefore, more information, especially regarding the chromosomal distribution of TIFY genes, is needed to accurately determine the evolutionary relatedness among these genes.
TIFY gene responses to drought conditions
Transcriptome data suggested that TIFY gene family members affect drought tolerance (Zhang et al. 2012). As determined by qRT-PCR analysis, the GaJAZ genes were more highly expressed in the three analyzed tissues than the other groups of TIFY genes. This indicates the JAZ subfamily might have a vital role in regulating drought tolerance in cotton. Therefore, we used GaJAZ5 in the drought assay.
When exposed to adverse environmental conditions, the stomata must close for plants to survive (Nauš et al. 2010). In this study, the stomatal apertures were smaller in the transgenic plants than in the WT controls. Stomatal closure reduces water loss, particularly in plants that have been exposed to water stress conditions caused by high solute concentrations (Wu et al. 2015) The observed reduced rate of water loss in transgenic plants might be caused by decreases in the size of the stomatal aperture. Additionally, compared with the WT plants, the GaJAZ5-expressing lines accumulated less H2O2, which may have protected cells from being damaged by drought or other stresses. During exposure to stress, increases in the metabolism of active oxygen species lead to the accumulation of H2O2 in plants. The accumulated H2O2 can directly or indirectly oxidize intracellular proteins, nucleic acids, and other biological macromolecules, as well as damage the cell membrane and accelerate cell collapse. Therefore, there is a close relationship between H2O2 content in plant tissues and plant responses to stress. The GaJAZ5-expressing lines responded better to drought conditions than the WT plants. The next step is to mimic the effects of drought through gene silencing or knockout.
Compared with the WT plants, the GaJAZ5-expressing lines were more drought resistant. Additionally, increased lateral root abundance and root hair density in GaJAZ5-expressing lines treated with methyl jasmonate can increase the total root surface area, which has implications for absorption of water and/or nutrients (Fig. S3). Following exposure to drought conditions or methyl jasmonate, JAZ proteins degrade and release transcription factors that can then increase the expression of downstream genes affecting drought resistance. The identities of the relevant G. arboreum transcription factors and target genes are currently unknown. Our future studies will focus on characterizing the mechanism of drought resistance and determining the relationship between drought stress and JA.
Conclusions
The TIFY gene family has been extensively studied in model plant species such as Arabidopsis, but there has been a lack of systematic analysis of TIFY family genes in cotton, especially in the A genome species G. arboreum. Here, we identified and compared the TIFY gene family members of the cotton species, G. raimondii and G. arboreum and G. hirsutum. Through evolutionary analysis, we explored the expansion and functional divergence of these TIFY genes. The TIFY genes likely experienced tandem and segmental duplication events, similar to the corresponding A. thaliana genes. Gene expression profiles revealed the GaJAZ genes have important functions during drought responses. Additionally, the results of the drought tolerance assays and qRT-PCR analyses of GaJAZ5 expression suggest that the JAZ subfamily may be involved in conferring drought tolerance to plants. Although, this study focused mainly on cotton TIFY genes, the relatively high conservation of orthologous TIFY family genes in various cotton interspecies indicates further comparative genomic analyses of other members of the family Malvaceae are warranted. These additional studies will help generate useful germplasm resources, which will be relevant for the breeding of drought-tolerant cotton species.
Abbreviations
- bHLH:
-
Basic helix-loop-helix
- PEG:
-
Polyethylene glycol
- JA:
-
Jasmonic acid
- SCFCOI1 :
-
Ubiquitin ligase complex
- ZIM:
-
Zinc finger protein expressed in inflorescence meristem
- dS :
-
Synonymous
- dN :
-
Non-synonymous
- JAZ:
-
Jasmonate ZIM domain
- Bp:
-
Base pair
- BLAST:
-
Basic local alignment search tool
- PCR:
-
Polymerase chain reaction
- qRT-PCR:
-
Quantitative reverse transcription PCR
- WT:
-
Wild-type
References
Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR (2004) The Pfam protein families database. Nucleic Acids Res 32:D138–D141
Berger S (2002) Jasmonate-related mutants of Arabidopsis as tools for studying stress signaling. Planta 214:497–504
Chen Y, Wang Y, Zhao T, Yang J, Feng S, Nazeer W, Zhang T, Zhou B (2015) A new synthetic amphiploid (AADDAA) between Gossypium hirsutum and G. arboreum lays the foundation for transferring resistances to verticillium and drought. PLoS One 10:e0128981
Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J (2009) Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. doi:10.1371/journal.pgen.1000440
Cheng Z, Sun L, Qi T, Zhang B, Peng W, Liu Y, Xie D (2011) The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol Plant 4:279–288
Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, Micol JL, Solano R (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448:666–671
Chothia C, Gough J, Vogel C, Teichmann SA (2003) Evolution of the protein repertoire. Science 300:1701–1703
Chung HS, Koo AJ, Gao X, Jayanty S, Thines B, Jones AD, Howe GA (2008) Regulation and function of Arabidopsis Jasmonate ZIM-domain genes in response to wounding and herbivory. Plant Physiol 146:952–964
Chung HS, Niu Y, Browse J, Howe GA (2009) Top hits in contemporary JAZ: an update on jasmonate signaling. Phytochemistry 70:1547–1559
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Conant GC, Wolfe KH (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9:938–950
Conconi A, Smerdon MJ, Howe GA, Ryan CA (1996) The octadecanoid signalling pathway in plants mediates a response to ultraviolet radiation. Nature 383:826–829
Conrad B, Antonarakis SE (2007) Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu Rev Genomics Hum Genet 8:17–35
Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48:355–381
d’Alencon E, Sezutsu H, Legeai F, Permal E, Bernard-Samain S, Gimenez S, Gagneur C, Cousserans F, Shimomura M, Brun-Barale A, Flutre T, Couloux A, East P, Gordon K, Mita K, Quesneville H, Fournier P, Feyereisen R (2010) Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements. Proc Natl Acad Sci USA 107:7680–7685
de Las Rivas B, Marcobal A, Munoz R (2004) Allelic diversity and population structure in Oenococcus oeni as determined from sequence analysis of housekeeping genes. Appl Environ Microbiol 70:7210–7219
Demianski AJ, Chung KM, Kunkel BN (2012) Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Mol Plant Pathol 13:46–57
Duan J, Zhang M, Zhang H, Xiong H, Liu P, Ali J, Li J, Li Z (2012) OsMIOX, a myo-inositol oxygenase gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). Plant Sci 196:143–151
Goldman N, Yang Z (1994) A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol 11:725–736
Grayson M (2013) Agriculture and drought. Nature 501:S1
Grunewald W, Vanholme B, Pauwels L, Plovie E, Inze D, Gheysen G, Goossens A (2009) Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Rep 10:923–928
Gu Z, Cavalcanti A, Chen FC, Bouman P, Li WH (2002) Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol 19:256–262
He H, Dong Q, Shao Y, Jiang H, Zhu S, Cheng B, Xiang Y (2012) Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep 31:1199–1217
He DH, Lei ZP, Tang BS, Xing HY, Zhao JX, Jing YL (2015) Identification and analysis of the TIFY gene family in Gossypium raimondii. Genet Mol Res 14:10119–10138
Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297
Kalivas A, Xanthopoulos F, Kehagia O, Tsaftaris AS (2011) Agronomic characterization, genetic diversity and association analysis of cotton cultivars using simple sequence repeat molecular markers. Genet Mol Res 10:208–217
Kumar R, Khurana A, Sharma AK (2014) Role of plant hormones and their interplay in development and ripening of fleshy fruits. J Exp Bot 65:4561–4575
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P (2006) SMART 5: domains in the context of genomes and networks. Nucleic Acids Res 34:D257–D260
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40:D302–D305
Li F, Fan G, Lu C, Xiao G, Zou C, Kohel RJ, Ma Z, Shang H, Ma X, Wu J, Liang X, Huang G, Percy RG, Liu K, Yang W, Chen W, Du X, Shi C, Yuan Y, Ye W, Liu X, Zhang X, Liu W, Wei H, Wei S, Huang G, Zhang X, Zhu S, Zhang H, Sun F, Wang X, Liang J, Wang J, He Q, Huang L, Wang J, Cui J, Song G, Wang K, Xu X, Yu JZ, Zhu Y, Yu S (2015) Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotech 33:524–530
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25:402–408
Lorenzo O, Chico JM, Sanchez-Serrano JJ, Solano R (2004) Jasmonate-insensitive1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16:1938–1950
McConn M, Browse J (1996) The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8:403
Mehetre S, Aher A, Gawande V, Patil V, Mokate A (2003) Induced polyploidy in Gossypium: a tool to overcome interspecific incompatibility of cultivated tetraploid and diploid cottons. Curr Sci 84:1510–1512
Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, Zeng W, Thines B, Staswick P, Browse J, Howe GA, He SY (2008) A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J 55:979–988
Moon J, Parry G, Estelle M (2004) The ubiquitin-proteasome pathway and plant development. Plant Cell 16:3181–3195
Nauš J (2010) Pareek, A., Sopory, S.K., Bohnert, H.J., Govindjee (ed.): abiotic stress adaptation in plants. Physiological, molecular and genomic foundation. Photosynthetica 48(3):474. doi:10.1007/s11099-010-0062-z
Nishii A, Takemura M, Fujita H, Shikata M, Yokota A, Kohchi T (2000) Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci Biotechnol Biochem 64:1402–1409
Ohno S, Wolf U, Atkin NB (1968) Evolution from fish to mammals by gene duplication. Hereditas 59:169–187
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23:1795–1814
Rao MV, Lee H, Creelman RA, Mullet JE, Davis KR (2000) Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12:1633–1646
Ren C, Han C, Peng W, Huang Y, Peng Z, Xiong X, Zhu Q, Gao B, Xie D (2009) A leaky mutation in DWARF4 reveals an antagonistic role of brassinosteroid in the inhibition of root growth by jasmonate in Arabidopsis. Plant Physiol 151:1412–1420
Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6:e230
Song S, Qi T, Wasternack C, Xie D (2014) Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr Opin Plant Biol 21:112–119
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612
Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J (2007) JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448:661–665
Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G (2007) The tify family previously known as ZIM. Trends Plant Sci 12:239–244
Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, Zou C, Li Q, Yuan Y, Lu C, Wei H, Gou C, Zheng Z, Yin Y, Zhang X, Liu K, Wang B, Song C, Shi N, Kohel RJ, Percy RG, Yu JZ, Zhu Y-X, Wang J, Yu S (2012) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44:1098–1103
Wang H, Liu G, Li C, Powell AL, Reid MS, Zhang Z, Jiang CZ (2013) Defence responses regulated by jasmonate and delayed senescence caused by ethylene receptor mutation contribute to the tolerance of petunia to Botrytis cinerea. Mol Plant Pathol 14:453–469
Wei K, Wang Y, Xie D (2014) Identification and expression profile analysis of the protein kinase gene superfamily in maize development. Mol Breeding 33:155–172
Wu T, Wang Y, Zheng Y, Fei Z, Dandekar AM, Xu K, Han Z, Cheng L (2015) Suppressing sorbitol synthesis substantially alters the global expression profile of stress response genes in apple (Malus domestica) leaves. Plant Cell Physiol 56:1748–1761
Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280:1091–1094
Yang S, Zhang X, Yue JX, Tian D, Chen JQ (2008) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomics 280:187–198
Ye H, Du H, Tang N, Li X, Xiong L (2009) Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol Biol 71:291–305
Zhang Y, Gao M, Singer SD, Fei Z, Wang H, Wang X (2012) Genome-wide identification and analysis of the TIFY gene family in grape. PLoS One 7:e44465
Zhang X, Yao D, Wang Q, Xu W, Wei Q, Wang C, Liu C, Zhang C, Yan H, Ling Y, Su Z, Li F (2013) mRNA-seq analysis of the Gossypium arboreum transcriptome reveals tissue selective signaling in response to water stress during seedling stage. PLoS One 8:e54762
Acknowledgments
We thank Dr. Jiahe Wu (Chinese Academy of Sciences, Beijing) and Dr. Maozhi Ren (Chongqing University, Chongqing) for comments on the manuscript. We also thank the Major Program of Joint Funds (Sinkiang) of the National Natural Science Foundation of China (Grant No. U1303282) and National Science Foundation (31501346) for funding this project.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
G. Zhao, Y. Song and C. Wang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Zhao, G., Song, Y., Wang, C. et al. Genome-wide identification and functional analysis of the TIFY gene family in response to drought in cotton. Mol Genet Genomics 291, 2173–2187 (2016). https://doi.org/10.1007/s00438-016-1248-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-016-1248-2