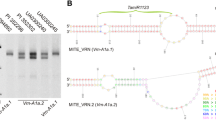
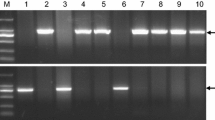
Abstract
Main conclusion
The durum wheat varieties from Ukraine, Russia, and Kazakhstan are characterized by the specific allelic composition of the VRN genes that sharply distinguish them from the Triticum durum varieties from other countries. For numerous varieties, the VRN alleles which previously were not found in tetraploid wheat were identified.
The ability of wheat to adapt to a wide range of environmental conditions is mostly determined by the allelic diversity within genes regulating the vernalization requirement (VRN) and photoperiod response (PPD). In the present study, allelic variation in the VRN1, VRN3, and PPD-A1 genes was investigated for 134 varieties of Triticum durum from different eco-geographic areas. It was shown that varieties from Russia and Ukraine have a specific allelic composition at the VRN genes, which in quantity and quality differed from European and American cultivars. A large number of varieties of T. durum from Russia carry the dominant Vrn-A1a.1 allele, previously identified mainly in hexaploid wheat. For some varieties from Eastern Europe and Asia, Vrn-A1i and vrn-A1b.3 recently revealed in wheat were also identified. Polymorphism of the VRN-B1 promoter region, distinguishing all three variants of this sequence (VRN-B1.f, VRN-B1.s, and VRN-B1.m), was detected. It was found that the dominant Vrn-B1c allele is commonly found in varieties of T. durum from Russia and Ukraine, but not Europe or USA. Furthermore, many Ukrainian and Russian varieties carry the dominant alleles of the both VRN-A1 and VRN-B1 genes simultaneously, while varieties from Europe and America carry the dominant allele of VRN-A1 alone. Finally, a high frequency of the Vrn-B3a allele, which previously was found only in some accessions of hexaploid wheat, was observed for varieties from Ukraine and Russia. It was revealed that the Ukrainian pool of T. durum varieties is currently the largest genetic source of the dominant Vrn-B3a allele in wheat in the worldwide.




Similar content being viewed by others
References
Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor Appl Genet 115(5):721–733
Chen F, Gao M, Zhang J, Zuo A, Shang X, Cui D (2013) Molecular characterization of vernalization and response genes in bread wheat from the Yellow and Huai Valley of China. BMC Plant Biol 13:199
Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F (2003) TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol 132(4):1849–1860
Derakhshan B, Mohammadi SA, Moghaddam M, Jalal Kamali MR (2013) Molecular characterization of vernalization genes in Iranian wheat landraces. Crop Breed J 3(1):1–11
Díaz A, Zikhali M, Turner AS, Isaac P, Laurie DA (2012) Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS One 7(3):e33234
Distelfeld A, Tranquilli G, Li C, Yan L, Dubcovsky J (2009) Genetic and molecular characterization of the VRN2 loci in tetraploid wheat. Plant Physiol 149(1):245–257
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Fu D, Szucs P, Yan L, Helguera M, Skinner JS, von Zitzewitz J, Hayes PM, Dubcovsky J (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genom 273(1):54–65
Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R (2007) Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinform 8(1):460–466
Iqbal M, Shahzad A, Ahmed I (2011) Allelic variation at the Vrn-A1, Vrn-B1, Vrn-D1, Vrn-B3 and Ppd-D1a loci of Pakistani spring wheat cultivars. Electron J Biotechnol 14(1):1–2
Kippes N, Debernardi JM, Vasquez-Gross HA, Akpinar BA, Budak H, Kato K, Chao S, Akhunov E, Dubcovsky J (2015) Identification of the VERNALIZATION 4 gene reveals the origin of spring growth habit in ancient wheats from South Asia. Proc Natl Acad Sci USA 112(39):E5401–E5410
Milec Z, Sumikova T, Tomkova L, Pankova K (2013) Distribution of different Vrn-B1 alleles in hexaploid spring wheat germplasm. Euphytica 192(3):371–378
Muterko AF, Balashova IA, Fayt VI, Sivolap YuM (2015a) Molecular genetic mechanisms of regulation of growth habit in wheat. Cytol Genet 49(1):58–71
Muterko A, Kalendar R, Cockram J, Balashova I (2015b) Discovery, evaluation and distribution of haplotypes and new alleles of the Photoperiod-A1 gene in wheat. Plant Mol Biol 88(1–2):149–164
Muterko A, Kalendar R, Salina E (2016) Novel alleles of the VERNALIZATION1 genes in wheat are associated with modulation of DNA curvature and flexibility in the promoter region. BMC Plant Biol 16(Suppl 1):9
Nishida H, Yoshida T, Kawakami K, Fujita M, Long B, Akashi Y, Laurie DA, Kato K (2013) Structural variation in the 5′ upstream region of photoperiod-insensitive alleles Ppd-A1a and Ppd-B1a identified in hexaploid wheat (Triticum aestivum L.), and their effect on heading time. Mol Breed 31(1):27–37
Shcherban AB, Efremova TT, Salina EA (2012) Identification of a new Vrn-B1 allele using two near-isogenic wheat lines with difference in heading time. Mol Breed 29(3):675–685
Takenaka S, Kawahara T (2012) Evolution and dispersal of emmer wheat (Triticum sp.) from novel haplotypes of Ppd-1 (photoperiod response) genes and their surrounding DNA sequences. Theor Appl Genet 125(5):999–1014
Wilhelm EP, Turner AS, Laurie DA (2009) Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.). Theor Appl Genet 118(2):285–294
Würschum T, Boeven PHG, Langer SM, Longin CFH, Leiser WL (2015) Multiply to conquer: copy number variations at Ppd-B1 and Vrn-A1 facilitate global adaptation in wheat. BMC Genet 16:96
Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100(10):6263–6268
Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J (2004a) Allelic variation at the VRN1 promoter region in polyploid wheat. Theor Appl Genet 109(8):1677–1686
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen J, Echenique V, Dubcovsky J (2004b) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303(5664):1640–1644
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103(51):19581–19586
Zhang XK, Xiao YG, Zhang Y, Xia XC, Dubcovsky J, He ZH (2008) Allelic variation at the vernalization genes Vrn-A1, Vrn-B1, Vrn-D1, and Vrn-B3 in Chinese wheat cultivars and their association with growth habit. Crop Sci 48(2):458–470
Acknowledgments
We are grateful to Carly Schramm for comments and English polishing of the manuscript. The analysis of 63 T. durum varieties (including all varieties from Russia) was supported by the Russian Science Foundation (Project No. 14-14-00161).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Muterko, A., Kalendar, R. & Salina, E. Allelic variation at the VERNALIZATION-A1, VRN-B1, VRN-B3, and PHOTOPERIOD-A1 genes in cultivars of Triticum durum Desf.. Planta 244, 1253–1263 (2016). https://doi.org/10.1007/s00425-016-2584-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2584-5