Abstract
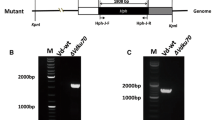
Normally, gene targeting by homologous recombination occurs rarely during a transformation process since non-homologous recombination is predominant in filamentous fungi. In our previous researches, the average gene replacement frequency (GRF) in Monascus ruber M7 was as low as 15 %. To develop a highly efficient gene targeting system for M. ruber M7, two M. ruber M7 null mutants of ku70 (MrΔku70) and ku80 (MrΔku80) were constructed which had no apparent defects in the development including vegetative growth, colony phenotype, microscopic morphology and spore yield compared with M. ruber M7. In addition, the production of some significant secondary metabolites such as pigments and citrinin had no differences between the two disruptants and the wild-type strain. Further results revealed that the GRFs of triA (encoding a putative acetyltransferase) were 42.2 % and 61.5 % in the MrΔku70 and MrΔku80 strains, respectively, while it was only about 20 % in M. ruber M7. Furthermore, GRFs of these two disruptants at other loci (the pigE, fmdS genes in MrΔku70 and the ku70 gene in MrΔku80) were investigated, and the results indicated that GRFs in the MrΔku70 strain and the MrΔku80 strain were doubled and tripled compared with that in M. ruber M7, respectively. Therefore, the ku70 and ku80 null mutants of M. ruber M7, especially the ku80-deleted strain, will be excellent hosts for efficient gene targeting.




Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aniya Y, Ohtani II, Higa T, Miyagi C, Gibo H, Shimabukuro M, Nakanishi H, Taira J (2000) Dimerumic acid as an antioxidant of the mold, Monascus anka. Free Radical Bio Med 28(6):999–1004. doi:10.1016/s0891-5849(00)00188-x
Asch DK, Kinsey JA (1990) Relationship of vector insert size to homologous integration during transformation of Neurospora crassa with the cloned am (GDH) gene. Mol Gen Genet 221(1):37–43
Aylon Y, Kupiec M (2004) DSB repair: the yeast paradigm. DNA Repair 3(8–9):797–815. doi:10.1016/j.dnarep.2004.04.013
Calsou P, Frit P, Humbert O, Muller C, Chen DJ, Salles B (1999) The DNA-dependent protein kinase catalytic activity regulates DNA end processing by means of Ku entry into DNA. J Biol Chem 274(12):7848–7856. doi:10.1074/jbc.274.12.7848
Carvalho NDSP, Arentshorst M, Kwon MJ, Meyer V, Ram AFJ (2010) Expanding the ku70 toolbox for filamentous fungi: establishment of complementation vectors and recipient strains for advanced gene analyses. Appl Microbiol Biotechnol 87(4):1463–1473
Cary RB, Peterson SR, Wang J (1997) DNA looping by Ku and the DNA-dependent protein kinase. Proc Natl Acad Sci USA 94(9):4267–4272. doi:10.1073/pnas.94.9.4267
Chang P-K, Scharfenstein LL, Wei Q, Bhatnagar D (2010) Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J Microbiol Meth 81(3):240–246. doi:10.1016/j.mimet.2010.03.010
Chen F, Hu X (2005) Study on red fermented rice with high concentration of monacolin K and low concentration of citrinin. Int J Food Microbiol 103(3):331–337. doi:10.1016/j.ijfoodmicro.2005.03.002
Chen Y-P, Tseng C-P, Liaw L-L, Wang C-L, Chen IC, Wu W-J, Wu M-D, Yuan G-F (2008) Cloning and characterization of Monacolin K biosynthetic gene cluster from Monascus pilosus. J Agr Food Chem 56(14):5639–5646. doi:10.1021/jf800595k
Chen Y-P, Yuan G-F, Hsieh S-Y, Lin Y-S, Wang W-Y, Liaw L-L, Tseng C-P (2009) Identification of the mokH gene encoding transcription factor for the upregulation of Monacolin K biosynthesis in Monascus pilosus. J Agr Food Chem 58(1):287–293. doi:10.1021/jf903139x
Chen W, Xie T, Shao Y, Chen F (2012a) Genomic characteristics comparisons of 12 food-related filamentous fungi in tRNA gene set, codon usage and amino acid composition. Gene 497(1):116–124. doi:10.1016/j.gene.2012.01.016
Chen W, Xie T, Shao Y, Chen F (2012b) Phylogenomic relationships between amylolytic enzymes from 85 strains of fungi. PLoS One 7(11):e49679. doi:10.1371/journal.pone.0049679
Choi YE, Shim WB (2008) Enhanced homologous recombination in Fusarium verticillioides by disruption of FvKU70, a gene required for a non-homologous end joining mechanism. Plant Pathol J 24(1):1–7
Choquer M, Robin G, Le Pêcheur P, Giraud C, Levis C, Viaud M (2008) Ku70 or Ku80 deficiencies in the fungus Botrytis cinerea facilitate targeting of genes that are hard to knock out in a wild-type context. FEMS Microbiol Lett 289(2):225–232. doi:10.1111/j.1574-6968.2008.01388.x
Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103(27):10352–10357. doi:10.1073/pnas.0601456103
Critchlow SE, Jackson SP (1998) DNA end-joining: from yeast to man. Trends Biochem Sci 23(10):394–398. doi:10.1016/s0968-0004(98)01284-5
da Silva Ferreira ME, Kress MRVZ, Savoldi M, Goldman MHS, Härtl A, Heinekamp T, Brakhage AA, Goldman GH (2006) The akuB KU80 mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell 5(1):207–211. doi:10.1128/ec.5.1.207-211.2006
Daley JM, Palmbos PL, Wu DL, Wilson TE (2005) Nonhomologous end joining in yeast. Annu Rev Genet 39:431–451
Doherty AJ, Jackson SP (2001) DNA repair: how Ku makes ends meet. Curr Biol 11(22):R920–R924. doi:10.1016/s0960-9822(01)00555-3
Dunlap JC, Borkovich KA, Henn MR, Turner GE, Sachs MS, Glass NL, McCluskey K, Plamann M, Galagan JE, Birren BW, Weiss RL, Townsend JP, Loros JJ, Nelson MA, Lambreghts R, Colot HV, Park G, Collopy P, Ringelberg C, Crew C, Litvinkova L, DeCaprio D, Hood HM, Curilla S, Shi M, Crawford M, Koerhsen M, Montgomery P, Larson L, Pearson M, Kasuga T, Tian C, Baştürkmen M, Altamirano L, Xu J (2007) Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv Genet 57:49–96
Dyck EV, Stasiak AZ, Stasiak A, West SC (1999) Binding of double-strand breaks in DNA by human Rad52 protein. Nature 398(6729):728–731
Fang Z, Zhang Y, Cai M, Zhang J, Zhang Y, Zhou X (2012) Improved gene targeting frequency in marine-derived filamentous fungus Aspergillus glaucus by disrupting ligD. J Appl Genet 53(3):355–362. doi:10.1007/s13353-012-0095-z
Feng J, Li W, Hwang S-F, Gossen BD, Strelkov SE (2012a) Enhanced gene replacement frequency in KU70 disruption strain of Stagonospora nodorum. Microbiol Res 167(3):173–178. doi:10.1016/j.micres.2011.05.004
Feng Y, Shao Y, Chen F (2012b) Monascus pigments. Appl Microbiol Biotechnol 96(6):1421–1440. doi:10.1007/s00253-012-4504-3
Goins CL, Gerik KJ, Lodge JK (2006) Improvements to gene deletion in the fungal pathogen Cryptococcus neoformans: absence of Ku proteins increases homologous recombination, and co-transformation of independent DNA molecules allows rapid complementation of deletion phenotypes. Fungal Genet Biol 43(8):531–544. doi:10.1016/j.fgb.2006.02.007
Gottlieb TM, Jackson SP (1993) The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72(1):131–142
Guangtao Z, Hartl L, Schuster A, Polak S, Schmoll M, Wang T, Seidl V, Seiboth B (2009) Gene targeting in a nonhomologous end joining deficient Hypocrea jecorina. J Biotechnol 139(2):146–151. doi:10.1016/j.jbiotec.2008.10.007
Haarmann T, Lorenz N, Tudzynski P (2008) Use of a nonhomologous end joining deficient strain (Δku70) of the ergot fungus Claviceps purpurea for identification of a nonribosomal peptide synthetase gene involved in ergotamine biosynthesis. Fungal Genet Biol 45(1):35–44. doi:10.1016/j.fgb.2007.04.008
Haber JE (2006) Transpositions and translocations induced by site-specific double-strand breaks in budding yeast. DNA Repair 5(9–10):998–1009
Hamid MI, Zeng F, Cheng J, Jiang D, Fu Y (2013) Disruption of heat shock factor 1 reduces the formation of conidia and thermotolerance in the mycoparasitic fungus Coniothyrium minitans. Fungal Genet Biol doi:http://dx.doi.org/10.1016/j.fgb.2012.12.002
Hoff B, Kamerewerd J, Sigl C, Zadra I, Kück U (2010) Homologous recombination in the antibiotic producer Penicillium chrysogenum: strain ΔPcku70 shows up-regulation of genes from the HOG pathway. Appl Microbiol Biotechnol 85(4):1081–1094. doi:10.1007/s00253-009-2168-4
Honda S, Selker EU (2009) Tools for fungal proteomics: multifunctional Neurospora vectors for gene replacement, protein expression and protein purification. Genetics 182(1):11–23. doi:10.1534/genetics.108.098707
Hood E, Gelvin S, Melchers L, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2(4):208–218. doi:10.1007/bf01977351
Hsu W-H, Pan T-M (2012) Monascus purpureus-fermented products and oral cancer: a review. Appl Microbiol Biotechnol 93(5):1831–1842. doi:10.1007/s00253-012-3891-9
Ishibashi K, Suzuki K, Ando Y, Takakura C, Inoue H (2006) Nonhomologous chromosomal integration of foreign DNA is completely dependent on MUS-53 (human Lig4 homolog) in Neurospora. Proc Natl Acad Sci USA 103(40):14871–14876. doi:10.1073/pnas.0604477103
Iwabuchi K, Hashimoto M, Matsui T, Kurihara T, Shimizu H, Adachi N, Ishiai M, Yamamoto K, Tauchi H, Takata M, Koyama H, Date T (2006) 53BP1 contributes to survival of cells irradiated with X-ray during G1 without Ku70 or Artemis. Genes Cells 11(8):935–948. doi:10.1111/j.1365-2443.2006.00989.x
Kanaar R, Hoeijmakers JHJ, van Gent DC (1998) Molecular mechanisms of DNA double-strand break repair. Trends Cell Biol 8(12):483–489. doi:10.1016/s0962-8924(98)01383-x
Kito H, Fujikawa T, Moriwaki A, Tomono A, Izawa M, Kamakura T, Ohashi M, Sato H, Abe K, Nishimura M (2008) MgLig4, a homolog of Neurospora crassa Mus-53 (DNA ligase IV), is involved in, but not essential for, non-homologous end-joining events in Magnaporthe grisea. Fungal Genet Biol 45(12):1543–1551. doi:10.1016/j.fgb.2008.09.005
Krappmann S, Sasse C, Braus GH (2006) Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot Cell 5(1):212–215
Kück U, Hoff B (2010) New tools for the genetic manipulation of filamentous fungi. Appl Microbiol Biotechnol 86(1):51–62. doi:10.1007/s00253-009-2416-7
Lai Y, Wang L, Qing L, Chen F (2011) Effects of cyclic AMP on development and secondary metabolites of Monascus ruber M-7. Lett Appl Microbiol 52(4):420–426. doi:10.1111/j.1472-765X.2011.03022.x
Larrondo LF, Colot HV, Baker CL, Loros JJ, Dunlap JC (2009) Fungal functional genomics: tunable knockout-knock-in expression and tagging strategies. Eukaryot Cell 8(5):800–804. doi:10.1128/ec.00072-09
Lee C-L, Pan T-M (2011) Red mold fermented products and Alzheimer's disease: a review. Appl Microbiol Biotechnol 91(3):461–469. doi:10.1007/s00253-011-3413-1
Lee B-H, Pan T-M (2012a) Benefit of Monascus-fermented products for hypertension prevention: a review. Appl Microbiol Biotechnol 94(5):1151–1161. doi:10.1007/s00253-012-4076-2
Lee C-L, Pan T-M (2012b) Development of Monascus fermentation technology for high hypolipidemic effect. Appl Microbiol Biotechnol 94(6):1449–1459. doi:10.1007/s00253-012-4083-3
Li L, Shao Y, Li Q, Yang S, Chen F (2010a) Identification of Mga1, a G-protein α-subunit gene involved in regulating citrinin and pigment production in Monascus ruber M7. FEMS Microbiol Lett 308(2):108–114. doi:10.1111/j.1574-6968.2010.01992.x
Li ZH, Du CM, Zhong YH, Wang TH (2010b) Development of a highly efficient gene targeting system allowing rapid genetic manipulations in Penicillium decumbens. Appl Microbiol Biotechnol 87(3):1065–1076. doi:10.1007/s00253-010-2566-7
Lin YL, Wang TH, Lee MH, Su NW (2008) Biologically active components and nutraceuticals in the Monascus-fermented rice: a review. Appl Microbiol Biotechnol 77(5):965–973. doi:10.1007/s00253-007-1256-6
Maruyama J-I, Kitamoto K (2008) Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (ΔligD) in Aspergillus oryzae. Biotechnol Lett 30(10):1811–1817. doi:10.1007/s10529-008-9763-9
Meyer V (2008) Genetic engineering of filamentous fungi—progress, obstacles and future trends. Biotechnol Adv 26(2):177–185. doi:10.1016/j.biotechadv.2007.12.001
Meyer V, Arentshorst M, El-Ghezal A, Drews A-C, Kooistra R, van den Hondel CAMJJ, Ram AFJ (2007) Highly efficient gene targeting in the Aspergillus niger kusA mutant. J Biotechnol 128(4):770–775. doi:10.1016/j.jbiotec.2006.12.021
Mizutani O, Kudo Y, Saito A, Matsuura T, Inoue H, Abe K, Gomi K (2008) A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet Biol 45(6):878–889. doi:10.1016/j.fgb.2007.12.010
Mladenov E, Iliakis G (2011) Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res 711(1–2):61–72. doi:10.1016/j.mrfmmm.2011.02.005
Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR (2006) A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172(3):1557–1566. doi:10.1534/genetics.105.052563
Ninomiya Y, Suzuki K, Ishii C, Inoue H (2004) Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci USA 101(33):12248–12253. doi:10.1073/pnas.0402780101
Paillard S, Strauss F (1991) Analysis of the mechanism of interaction of simian Ku protein with DNA. Nucleic Acids Res 19(20):5619–5624. doi:10.1093/nar/19.20.5619
Pastink A, Eeken JCJ, Lohman PHM (2001) Genomic integrity and the repair of double-strand DNA breaks. Mutat Res 480–481:37–50. doi:10.1016/s0027-5107(01)00167-1
Pattanagul P, Pinthong R, Phianmongkhol A, Tharatha S (2008) Mevinolin, citrinin and pigments of adlay angkak fermented by Monascus sp. Int J Food Microbiol 126(1–2):20–23. doi:10.1016/j.ijfoodmicro.2008.04.019
Pöggeler S, Kück U (2006) Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene 378(0):1–10. doi:10.1016/j.gene.2006.03.020
Ramsden DA, Gellert M (1998) Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J 17(2):609–614
Shao YC, Ding YD, Zhao Y, Yang S, Xie BJ, Chen FS (2009) Characteristic analysis of transformants in T-DNA mutation library of Monascus ruber. World J Microb Biot 25(6):989–995. doi:10.1007/s11274-009-9977-6
Shi YC, Pan TM (2011) Beneficial effects of Monascus purpureus NTU 568-fermented products: a review. Appl Microbiol Biotechnol 90(4):1207–1217. doi:10.1007/s00253-011-3202-x
Shi Y-C, Pan T-M (2012) Red mold, diabetes, and oxidative stress: a review. Appl Microbiol Biotechnol 94(1):47–55. doi:10.1007/s00253-012-3957-8
Shrivastav M, De Haro LP, Nickoloff JA (2008) Regulation of DNA double-strand break repair pathway choice. Cell Res 18(1):134–147
Su Y-C, Wang J-J, Lin T-T, Pan T-M (2003) Production of the secondary metabolites ϒ-aminobutyric acid and monacolin K by Monascus. J Ind Microbiol Biot 30(1):41–46
Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR (2007) Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc 1(6):3111–3120
Tachibana A (2004) Genetic and physiological regulation of non-homologous end-joining in mammalian cells. Adv Biophys 38:21–44
Takahashi T, Masuda T, Koyama Y (2006a) Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol Genet Genom 275:460–470. doi:10.1007/s00438-006-0104-1
Takahashi T, Masuda T, Koyama Y (2006b) Identification and analysis of Ku70 and Ku80 homologs in the koji molds Aspergillus sojae and Aspergillus oryzae. Biosci Biotechnol Biochem 70(1):135–143
Takahashi T, Jin FJ, Sunagawa M, Machida M, Koyama Y (2008) Generation of large chromosomal deletions in koji molds Aspergillus oryzae and Aspergillus sojae via a loop-out recombination. Appl Environ Microb 74(24):7684–7693. doi:10.1128/aem.00692-08
Thompson LH (2012) Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat Res-Rev Mut 751(2):158–246. doi:10.1016/j.mrrev.2012.06.002
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680. doi:10.1093/nar/22.22.4673
Verbeke J, Beopoulos A, Nicaud J-M (2012) Efficient homologous recombination with short length flanking fragments in Ku70 deficient Yarrowia lipolytica strains. Biotechnol Lett:1–6 doi:10.1007/s10529-012-1107-0
Villalba F, Collemare J, Landraud P, Lambou K, Brozek V, Cirer B, Morin D, Bruel C, Beffa R, Lebrun M-H (2008) Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genet Biol 45(1):68–75. doi:10.1016/j.fgb.2007.06.006
Walker JR, Corpina RA, Goldberg J (2001) Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature 412(6847):607–614, http://www.nature.com/nature/journal/v412/n6847/suppinfo/412607a0_S1.html
Wang H, Perrault AR, Takeda Y, Qin W, Iliakis G (2003) Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res 31(18):5377–5388
Weld RJ, Plummer KM, Carpenter MA, Ridgway HJ (2006) Approaches to functional genomics in filamentous fungi. Cell Res 16(1):31–44. doi:10.1038/sj.cr.7310006
Weterings E, van Gent DC (2004) The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair 3(11):1425–1435. doi:10.1016/j.dnarep.2004.06.003
Yang Y, Li L, Li X, Shao Y, Chen F (2012) mrflbA, encoding a putative FlbA, is involved in aerial hyphal development and secondary metabolite production in Monascus ruber M-7. Fungal Biol 116(2):225–233. doi:10.1016/j.funbio.2011.11.005
Yasuda M, Tachibana S, Kuba-Miyara M (2012) Biochemical aspects of red koji and tofuyo prepared using Monascus fungi. Appl Microbiol Biotechnol 96(1):49–60. doi:10.1007/s00253-012-4300-0
Yu Y, Du J, Wang G, Ji J (2003) Studies on the freeze-thaw method of transforming recombinant plasmid DNA into Agrobacterium tumefaciens. J Jilin Agricultural University 25:257–259 (in Chinese)
Yu J-H, Hamari Z, Han K-H, Seo J-A, Reyes-Domínguez Y, Scazzocchio C (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41(11):973–981. doi:10.1016/j.fgb.2004.08.001
Acknowledgements
We thank Dr. Daohong Jiang from Plant Pathology, College of Plant Science and Technology, Huazhong Agricultural University for providing plasmids pKN1 and pSKH. This work was financially supported by the programs of National Natural Science Foundation of China (No.31171649; No.31271834), New Century of Chinese Ministry of Education (NCET-05-0667) and National High Technology Research and Development Program of China (2006AA10Z1A3).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
He, Y., Liu, Q., Shao, Y. et al. ku70 and ku80 null mutants improve the gene targeting frequency in Monascus ruber M7. Appl Microbiol Biotechnol 97, 4965–4976 (2013). https://doi.org/10.1007/s00253-013-4851-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-4851-8