Abstract
Aims/hypothesis
Existing thiazolidinedione (TZD) drugs for diabetes have severe side effects. The aim of this study is to develop alternative peroxisome proliferator-activated receptor γ (PPARγ) ligands that retain the benefits in improving insulin resistance but with reduced side effects.
Methods
We used AlphaScreen assay to screen for new PPARγ ligands from compound libraries. In vitro biochemical binding affinity assay and in vivo cell-based reporter assay were used to validate ionomycin as a partial ligand of PPARγ. A mouse model of diabetes was used to assess the effects of ionomycin in improving insulin sensitivity. Crystal structure of PPARγ complexed with ionomycin revealed the unique binding mode of ionomycin, which elucidated the molecular mechanisms allowing the discrimination of ionomycin from TZDs.
Results
We found that the antibiotic ionomycin is a novel modulating ligand for PPARγ. Both the transactivation and binding activity of PPARγ by ionomycin can be blocked by PPARγ specific antagonist GW9662. Ionomycin interacts with the PPARγ ligand-binding domain in a unique binding mode with properties and epitopes distinct from those of TZD drugs. Ionomycin treatment effectively improved hyperglycaemia and insulin resistance, but had reduced side effects compared with TZDs in the mouse model of diabetes. In addition, ionomycin effectively blocked the phosphorylation of PPARγ at Ser273 by cyclin-dependent kinase 5 both in vitro and in vivo.
Conclusions/interpretation
Our studies suggest that ionomycin may represent a unique template for designing novel PPARγ ligands with advantages over current TZD drugs.
Similar content being viewed by others
Introduction
Nuclear receptors (NRs) are crucial transcriptional factors that control gene expression by binding to specific response elements of their target genes [1, 2]. Small molecules that act as ligands for NRs can enhance or reduce the transcriptional activity of NRs by recruiting coactivators or corepressors. Peroxisome proliferator-activated receptors (PPARs) α, β and γ play a critical role in the physiological and pathological regulation of various biological activities, including metabolism, adipocyte differentiation, bone morphogenesis, inflammation and atherosclerosis [3–6]. The most prominent role of PPARγ is its contribution in improving insulin sensitivity upon binding to specific ligands [7–9]. Among PPARγ ligands, thiazolidinedione (TZDs) can alter the expression of PPARγ target genes, which are necessary for proper insulin sensitivity and have an impact on peripheral tissue insulin sensitivity, so that they are called insulin sensitisers. However, TZDs also display severe adverse effects, giving rise to fluid retention, weight gain, liver toxicity and cardiovascular disease, which are prevalent among diabetic patients [10–12]. Therefore, it is imperative to develop alternative PPARγ ligands that retain the benefits in improving insulin resistance but that have reduced side effects.
As a key structural feature of all NRs, the ligand-binding pocket of the PPARγ ligand-binding domain (LBD) includes an activation function 2 motif that displays great flexibility in response to diverse ligands, resulting in the transcriptional regulation of downstream PPARγ target genes [13–15]. These genes in turn regulate many metabolic pathways involved in glucose homeostasis and insulin sensitivity. Interestingly, some selective PPARγ modulators (SPPARMs) with partial or no agonism in transcriptional activity, have shown similar glucose-lowering effects to rosiglitazone but with reduced side effects, suggesting that PPARγ transcriptional activity may not be directly associated with insulin sensitivity [16–18]. Recently, the phosphorylation of PPARγ by cyclin-dependent kinase 5 (CDK5) was revealed as a critical link between PPARγ ligands and their glucose-lowering effects, thus providing a new avenue for the mechanistic understanding of PPARγ ligands in improving insulin sensitivity, and further uncovering a drug design strategy targeting PPARγ [19, 20]. Independent of transcriptional agonism, the CDK5-mediated phosphorylation of PPARγ at serine 273 plays a key role in insulin resistance and obesity. Obesity induced CDK5 cofactor p35 to truncate into p25, which can stimulate activation of CDK5 to phosphorylate PPARγ at Ser273, whereas the ligand binding of PPARγ reduced the binding of PPARγ to NR corepressor (NCoR) and CDK5, resulting in the decreased phosphorylation of PPARγ [21], which correlates well with the glucose-lowering effects.
Here, we sought to find alternative PPARγ ligands that retain the benefits in improving insulin resistance but with reduced side effects, and to further provide new drug design strategies for future pharmacological agents targeting PPARγ.
Methods
Protein preparation
Human PPARγ LBD (residues 206–477) was expressed as an N-terminal 6×His fusion protein from the expression vector pET24a (Novagen, Darmstadt, Germany). BL21 (DE3) cells transformed with the expression plasmid were grown in LB broth (Luria Broth Base) at 25°C to an optical density at 600 nm (OD600) of ∼1.0 and induced with 0.1 mmol/l isopropyl 1-thio-β-d-galactopyranoside (IPTG) at 16°C. The PPARγ LBD was purified with a 5 ml HiTrap HP column chelated with NiSO4, followed by a Q-Sepharose column (GE Healthcare, Pittsburgh, PA, USA). To prepare the protein–ligand complex, a fivefold excess of the ionomycin free acid (Sigma, St Louis, MO, USA) was added to the purified protein, followed by filter concentration to 10 mg/ml. The PPARγ LBD was complexed with twofold of steroid receptor coactivator 1 (SRC1) peptide (SLTERHKI LHRLLQEGSP) before filter concentration.
LanthaScreen
LanthaScreen TR-FRET PPARγ competitive binding assay was performed according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). A serial concentration of ionomycin or rosiglitazone (Sigma) (dissolved in DMSO and diluted in TR-FRET assay buffer, final concentration of DMSO is 1% vol./vol.) were put in a 384-well opti-plate (Perkin Elmer, Norwalk, CT, USA). A mixture of 5 nmol/l glutathione S-transferase (GST)-fused PPARγ LBD, 5 nmol/l Tb-GST-antibody and 5 nmol/l Fluormone Pan-PPAR Green were added to each sample well to a total volume of 40 μl. DMSO at 1% (vol./vol.) final concentration was used as a no-ligand control. The assay was performed in triplicate and the mixture was incubated for 2 h in the dark before analysis. The FRET signal was measured by excitation at 340 nm and emission at 520 nm for fluorescein and 495 nm for terbium. The fold change over DMSO was calculated by the ratio of 520 nm/495 nm. Graphs were plotted as fold change of FRET signal for each compound over DMSO-only control.
Crystallisation, data collection and structure determination
Crystals of PPARγ/ionomycin complex were grown at room temperature in hanging drops containing 1.0 μl of the above protein–ligand solutions and 1.0 μl of well buffer containing 0.2 mol/l sodium acetate, 5% (vol./vol.) ethylene glycol and 20% (wt/vol.) PEG 3350. The crystals were directly flash frozen in liquid nitrogen for data collection. The observed reflections were reduced, merged and scaled with DENZO and SCALEPACK in the HKL2000 package [22]. The structures were determined by molecular replacement in the CCP4 suite (www.ccp4.ac.uk). Manual model building was carried out with Coot [23], followed by REFMAC refinement in the CCP4 suite.
Cofactor binding assays
The binding of the various coregulator peptide motifs to PPARγ LBD in response to ligands was determined by AlphaScreen assays using a hexahistidine detection kit from Perkin Elmer as described previously [24]. The experiments were conducted with approximately 20–40 nmol/l receptor LBD and 20 nmol/l biotinylated coregulator peptides in the presence of 5 μg/ml donor and acceptor beads in a buffer containing 50 mmol/l 3-(N-morpholino)propanesulfonic acid (MOPS), 50 mmol/l NaF, 0.05 mmol/l N-cyclohexyl-3-aminopropanesulfonic acid (CHAPS) and 0.1 mg/ml bovine serum albumin, all adjusted to pH 7.4. The peptides with an N-terminal biotinylation are listed in electronic supplementary material (ESM) Table 1.
Transient transfection assay
Cos7 cells (ATCC, Manassas, VA, USA) were maintained in DMEM containing 10% (wt/vol.) fetal bovine serum and were transiently transfected using Lipofectamine 2000 (Invitrogen) [24]. All mutant PPARγ plasmids were created using the Quick-Change site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). Twenty-four-well plates were plated 24 h before transfection (5 × 104 cells per well). For Gal4-driven reporter assays, the cells were transfected with 200 ng Gal4-LBDs of various NRs and 200 ng of pG5Luc reporter (Promega, Madison, WI, USA). For native promoter reporter assays, the cells were cotransfected with plasmids encoding full-length PPARγ and peroxisome proliferator hormone response element (PPRE). Ligands were added 5 h after transfection. Cells were harvested 24 h later for the luciferase assays. Luciferase activity was normalised to renilla activity cotransfected as an internal control.
In vitro kinase assay
In vitro CDK assay was performed as previously described [19]. Briefly, 1 μg of purified His-tagged PPARγ LBD (residues 206–477) was incubated with 50 ng active CDK5/p25 (Invitrogen) in assay buffer (25 mmol/l Tris pH 7.5, 10 mmol/l MgCl2, 5 mmol/l β-glycerophosphate, 0.1 mmol/l Na3VO4, 2 mmol/l dithiothreitol [DTT]) (Cell Signaling Technology, Danvers, MA, USA) containing 100 μmol/l ATP in a 50-μl reaction volume for 30 min at room temperature. PPARγ ligands were pre-incubated with PPARγ LBD protein for 30 min before the assay was performed. Phosphorylation of PPARγ LBD was analysed by western blotting with anti-CDK substrate antibody (Cell Signaling Technology).
Preparation of white adipose tissue lysates
White adipose tissues from mice treated with compounds were homogenised in RIPA buffer (50 mmol/l Tris pH 7.5, 150 mmol/l NaCl, 1% (vol./vol.) NP-40, 0.5% (wt/vol.) sodium deoxycholate, 0.1% (wt/vol.) SDS with protease and phosphatase inhibitors). For western blotting, a rabbit polyclonal phospho-specific antibody against PPARγ Ser273 was produced by AbMax Biotechnology (Shanghai, China), with a synthetic phosphopeptide as previously described [19]. Total tissue lysates were analysed with anti-PPARγ antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Animal experiments
Animal experiments were performed according to procedures approved by the Institutional Animal Use and Care Committee of Xiamen University, China. Male mice (db/db and KK-Ay mice; Hua Fukang, Beijing, China), 8–10 weeks old, were acclimatised for 7 days under standard conditions before experiments. Mice were fed with a high-fat diet (D12492; Research Diets, New Brunswick, NJ, USA) and 5 mg/kg of rosiglitazone or 3 mg/kg of ionomycin were administered by i.p. injection once daily with vehicle (40% (wt/vol.) of 2-hydroxypropyl-β-cyclodextrin [HBC]) (Sigma) for 14 days. Mice were killed after 6 h of fasting, and serum samples were collected to measure the glucose and insulin levels.
Glucose and insulin tolerance, glucose and insulin levels
Mice treated with drugs were fasted for 6 h with free access to water. For the glucose tolerance test (GTT), 1 g/kg of glucose was injected intraperitoneally and blood glucose was measured with the Accu-Check Performa (Roche Applied Science, Mannheim, Germany ) at 0, 30, 60, 90 and 120 min. For the insulin tolerance test (ITT), 1 U/kg of recombinant human insulin (Novolin 30R; Novo Nordisk, Bagsvaerd, Denmark) was injected intraperitoneally, and blood glucose was measured at 0, 30, 60, 90 and 120 min after insulin injection. Serum glucose levels were determined by the Glucose Oxidase Method (Applygen, Beijing, China) and serum insulin levels were determined by ELISA using an ultra-sensitive mouse insulin kit (Crystal Chem, Downers Grove, IL, USA).
Gene expression analysis
Total RNA was isolated from epididymal fat pads using a Tissue RNA kit (Omega Bio-Tek, Norcross, GA, USA). The RNA was reverse-transcribed using the reverse transcription kit (Takara, Dalian, China). Quantitative PCR reactions were performed with SYBR green fluorescent dye using a CFXTM96 real-time system (Bio-Rad, Hercules, CA, USA). Relative mRNA expression was determined by the ΔΔCt method (www.bio-rad.com/amplification/) normalised to actin levels. The sequences of primers are listed in ESM Table 2.
Results
Identification of ionomycin as a novel PPARγ ligand
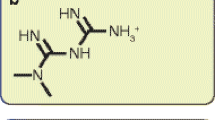
To search for novel ligands for PPARγ, we used PPARγ LBD as a bait to screen chemical libraries based on AlphaScreen biochemical assay, which determines the efficacy of small molecules in influencing binding affinity of PPARγ with coregulator peptides [25–27]. Surprisingly an antibiotic, ionomycin, was unravelled as a positive PPARγ activator from the Enzo Rare Natural Products Library, which contains 82 purified compounds from natural sources. Notably, the chemical structure of ionomycin shows a molecular scaffold distinct from that of TZDs (Fig. 1a). To confirm the specificity of ionomycin in activating PPARγ, Cos7 cells were cotransfected with a Gal4-driven reporter together with plasmids encoding various NR LBDs fused with the Gal4 DNA-binding domain. In agreement with the initial AlphaScreen biochemical assay, treatment with ionomycin significantly induced the transcriptional activity of PPARγ, to a lesser extent than rosiglitazone, but with no impact on a variety of other NRs tested (Fig. 1b). LanthaScreen TR-FRET competitive binding assay showed that ionomycin had a half-maximum inhibitory concentration (IC50) of about 500 nmol/l (Fig. 1c) to PPARγ, which is tenfold less than that of rosiglitazone but similar to that of other TZDs, such as pioglitazone, and many SPPARMs [17, 18]. Our results indicate that ionomycin selectively recognises and interacts with PPARγ LBD among various NRs.
Identification of ionomycin as a PPARγ partial agonist. (a) Chemical structures of ionomycin and rosiglitazone. (b) Receptor-specific transactivation by ionomycin. Cos7 cells were cotransfected with pG5Luc reporter together with the plasmids encoding various NR LBDs fused with the Gal4 DNA-binding domain. After transfection, cells were treated with DMSO (white bars), 1 μmol/l ionomycin (black bars) or ligands specific for each receptor (striped bars): PPARγ, 1 μmol/l rosiglitazone; PPARα, 1 μmol/l GW590735; PPARδ, 1 μmol/l GW0472; GR, 0.1 μmol/l dexamethasone; LXRα, 1 μmol/l T0901317. Luciferase activity is reported normalised to renilla activity, cotransfected as an internal control. ERR, oestrogen (estrogen)-related receptor; GR, glucocorticoid receptor; LXR, liver X receptor; ROR, retinoid-related orphan receptor; RTR, retinoid receptor-related testis-associated receptor; TR3, nuclear receptor subfamily 4 group A member 1 (NR4A1). (c) The binding affinity of ionomycin to PPARγ by LanthaScreen assay. Triangles, rosiglitazone; squares, ionomycin. Values are the means±SEM of three independent experiments
To further examine the biochemical mechanism of PPARγ activation by ionomycin, we determined the ability of ionomycin to promote the recruitment of coregulator motifs by PPARγ using AlphaScreen biochemical assay. As shown in Fig. 2a, ionomycin enhanced the interaction of PPARγ with various coactivator LXXLL motifs from the family of SRCs (SRC1-2, SRC2-3 and SRC3-3), Trap220 and PGC-1α, but decreased the binding of a corepressor motif from NCoR (NCoR-2) with PPARγ. Ionomycin displayed a weaker effect than rosiglitazone in recruiting coactivators, suggesting a partial agonist nature, which is consistent with the results from reporter assays. Similar to rosiglitazone, ionomycin also promoted the interaction of PPARγ with coactivator motifs in a concentration-dependent manner, with tenfold less potency than rosiglitazone (Fig. 2b). GW9662 is an antagonist for PPARγ that covalently occupies the same binding pocket as agonists and irreversibly antagonises their binding to PPARγ [28]. As shown in Fig. 3 a–d, results from AlphaScreen indicate that GW9662 blocks the coactivator recruitment of PPARγ induced by both rosiglitazone and ionomycin. Further, ionomycin-mediated transcriptional activity of PPARγ was also inhibited by the PPARγ-specific antagonist GW9662 in the cell-based reporter assays (Fig. 3e), reaffirming that the effect of ionomycin on PPARγ functions through direct binding to this receptor.
Binding of various cofactor LXXLL motifs to PPARγ LBD using AlphaScreen assays. (a) Binding of cofactor binding motifs to PPARγ LBD in response to 1 μmol/l rosiglitazone or 5 μmol/l ionomycin. Background reading with the PPARγ LBD is less than 300 photon counts. White bars, DMSO; striped bars, rosiglitazone; black bars, ionomycin. (b) The relative cofactor binding affinity (compared with maximum activity) of coactivator LXXLL motif SRC2-3 to PPARγ LBD is dose dependent on rosiglitazone (triangles) and ionomycin (squares) treatment. The peptide sequences are listed in the Methods section. Values are the means±SEM of three independent experiments
Ionomycin functions as a partial agonist through direct interaction with PPARγ. (a–d) The binding of PPARγ LBD with various coactivator LXXLL motifs in response to ionomycin was blocked by PPARγ antagonist GW9662 in AlphaScreen assays. Both ionomycin and GW9662 were used at a concentration of 1 μmol/l. (e) The transcriptional activity of PPARγ induced by ionomycin was inhibited by PPARγ antagonist GW9662. Cos7 cells were cotransfected with plasmids encoding full-length PPARγ and a PPRE luciferase reporter. After transfection, cells were treated with ionomycin or rosiglitazone together with various concentrations of GW9662. White bars, DMSO; striped bars, rosiglitazone; black bars, ionomycin. Values are the means±SEM of three independent experiments
Ionomycin effectively improved hyperglycaemia in a mouse model of diabetes, with reduced side effects
Since ionomycin displays typical features of a partial agonist of PPARγ, we were curious to know whether ionomycin had effects on diabetes like those of other PPARγ ligands or modulators. We used db/db mice to investigate the effect of ionomycin in vivo. Neither ionomycin nor rosiglitazone had a significant effect on food intake (Fig. 4a). Similar to rosiglitazone, ionomycin significantly reduced the levels of serum glucose and insulin (Fig. 4 c, d). To further investigate whether ionomycin could improve glucose tolerance and insulin sensitivity, we performed a GTT and an ITT. Both ionomycin- and rosiglitazone-injected mice showed improved insulin sensitivity and glucose tolerance compared with vehicle-treated controls (Fig. 4 e, f).
Ionomycin improved glucose tolerance and insulin sensitivity with reduced adipogenesis activity in db/db mice. The food intake (a), body weight (b), serum glucose (c) and insulin (d) levels, and the weight of epididymal fat pads (Epi-WAT) (g) were determined after db/db mice were injected intraperitoneally with vehicle (HBC, white bars or diamond symbols), rosiglitazone (striped bars or square symbols) or ionomycin (black bars or triangle symbols) (n = 4 for each group) for 14 days. Insulin (1 U/kg) and glucose (1 g/kg), respectively, were administered by i.p. injection to 6-h-fasted db/db mice (n = 4 for each group) for ITT (e) and GTT (f). (h) The mRNA levels of PPARγ target genes as well as genes involved in adipogenesis in epididymal fat pads. Error bars are SEM; *p < 0.05, **p < 0.01 compared with vehicle
Remarkably, the body weight of ionomycin-treated db/db mice did not change compared with vehicle control, while the rosiglitazone-treated mice showed significant weight gain (Fig. 4b), which is consistent with the reported side effects associated with TZD drugs [29, 30]. Further, the weight of epididymal fat pads from ionomycin-treated mice was similar to that of vehicle controls, significantly less than that of rosiglitazone-treated mice (Fig. 4g), which is consistent with the results in 3T3-L1 cells (ESM Fig. 1). In addition, we also detected the expression of genes in adipose tissues, including PPARγ target genes Ap2 and Cd36, and genes involved in adipogenesis, such as CCAAT-enhancer binding protein (C/ebpα [also known as Cebpa]) gene, glucose transporter (Glut4), sterol regulatory element-binding protein-1c gene (Srebp-1c [also known as Srebf1]), acetyl-CoA carboxylase (Acc [also known as Acaca]), fatty acid synthase (Fasn) gene and stearoyl-CoA desaturase-1 (Scd1). As expected, ionomycin did not induce adipogenic genes compared with control (Fig. 4h). In contrast, the mRNA levels of most of these genes were significantly increased in adipose tissues of rosiglitazone-treated mice, which is consistent with the published results [29, 30]. Moreover, genes encoding adiponectin (Adipoq) and adipsin (Cfd), dysregulated in obesity, were also elevated by the ionomycin treatment (Fig. 4h). To further test the effects of ionomycin in other diabetic models, we performed similar experiments in KK-Ay mice. All the results are consistent with those from db/db mice (ESM Fig. 2), indicating that ionomycin functions as an insulin sensitiser in mice with various genetic backgrounds.
To detect the effects of ionomycin on fluid retention, the mRNA levels of genes such as epithelial sodium channel γ (EnaCγ [also known as Scnn1g]), aquaporin-2 (Aqp2) and serum/glucocorticoid regulated kinase 1 (Sgk1) were analysed in compound-treated mouse renal inner medullary collecting duct (mIMCD-3) cells. Unlike rosiglitazone, ionomycin treatment elicited either significant suppression or no induction of the mRNA levels of genes involved in fluid accumulation in mIMCD-3 cells (ESM Fig. 3).
To assess the hepatotoxicity of ionomycin, we analysed several specific aspects related to hepatotoxicity. As shown in ESM Fig. 4a, treatment of db/db mice with ionomycin downregulated inflammation-related genes, such as Cxcl10 and Ifnγ (also known as Ifng), which is beneficial for liver protection. Further, the ratio of liver/body weight was significantly reduced by the ionomycin treatment (ESM Fig. 4b). Consistently, the triacylglycerol level in the liver tissue was also significantly lower in ionomycin-treated mice than in those treated by rosiglitazone (ESM Fig. 4c). These findings were further confirmed by the regulation of genes involved in fat synthesis (ESM Fig. 4d). Moreover, similar to rosiglitazone, ionomycin had a minimal effect on the viability of HepG2 and mIMCD-3 cells as determined by MTT assay (ESM Fig. 4e). These results suggest that ionomycin has reduced side effects for some physiological variables compared with TZDs.
Ionomycin effectively blocked the CDK5-mediated phosphorylation of PPARγ at Ser273 in vitro and in vivo
Choi et al have shown that PPARγ ligands (including agonists such as rosiglitazone and SPPARMs such as MRL24) blocked the obesity-linked phosphorylation of PPARγ by CDK5 at Ser273, and this inhibition of phosphorylation proved to be tightly correlated with the glucose-lowering effects of the PPARγ ligands in obese mice and diabetic patients [19]. As such, we performed an in vitro CDK kinase assay to test whether ionomycin has this biochemical function. Interestingly, although having lower PPARγ transcriptional activity, ionomycin effectively blocked the CDK5-mediated phosphorylation of PPARγ with similar potency to rosiglitazone (Fig. 5a). Consistently, the data from the mammalian two hybrid assay indicated that ionomycin and rosiglitazone caused a similar reduction in binding of NCoR to PPARγ (ESM Fig. 5), which is critical for the PPARγ phosphorylation by CDK5 [21]. To test whether ionomycin blocked the phosphorylation of PPARγ at Ser273 in vivo, db/db mice were injected intraperitoneally with vehicle, rosiglitazone or ionomycin once a day for 14 days. Consistent with the data in vitro, both rosiglitazone and ionomycin caused a similar reduction in PPARγ phosphorylation at Ser273 in white adipose tissues (Fig. 5b, c), suggesting that ionomycin may exert an anti-diabetic effect through modulating CDK5-dependent PPARγ phosphorylation.
Ionomycin blocked the phosphorylation of PPARγ at Ser273 mediated by CDK5. (a) In vitro CDK5 assay on PPARγ LBD incubated with rosiglitazone or ionomycin. (b) Phosphorylation of PPARγ in white adipose tissue (WAT). (c) Quantification of PPARγ phosphorylation compared with total PPARγ. IB, immunoblot; Iono, ionomycin; pPPARγ, phosphorylated PPARγ; pSer273, phosphorylated PPARγ at Ser273; Rosi, rosiglitazone. Total WAT lysates were obtained from compound-treated db/db mice (n = 4 for each group), and immunoblotting was performed. Error bars are SEM; **p < 0.01 compared with vehicle
PPARγ recognises ionomycin with a unique binding mode
As a partial agonist of PPARγ with a weaker transcriptional activity, ionomycin has a distinct scaffold with a much larger size than that of the full agonist rosiglitazone (Fig. 1a), thus we wanted to elucidate the structural basis for the recognition of this unique ligand by PPARγ. To determine the binding characteristics of ionomycin to PPARγ, we solved the crystal structure of PPARγ complexed with ionomycin and the SRC1-2 LXXLL motif at 2.5 Å resolution (ESM Table 3). The structure reveals that the ionomycin-bound PPARγ LBD adopts a classical structure of a three-layer antiparallel α-helical sandwich, which is largely conserved among NR LBDs (Fig. 6a) [31, 32]. Inside the ligand-binding pocket of PPARγ, the existence of ionomycin was apparent from the highly revealing electron density map (Fig. 6b). Alignment of structures of PPARγ/ionomycin and PPARγ/rosiglitazone revealed that both ligand-bound PPARγ LBDs aligned well with ligands occupying the similar binding site in the PPARγ pocket (Fig. 6c, d).
The structure of the PPARγ LBD complexed with ionomycin. (a) The structures of PPARγ LBD bound with ionomycin in ribbon representation. PPARγ LBD is shown in red and the SRC1 motif is in yellow. Ionomycin is shown in stick representation with carbon and oxygen atoms depicted in green and red, respectively. (b) 2Fo-Fc electron density map (1.0σ) showing ionomycin bound to the PPARγ LBD. (c) Alignment of the PPARγ–ionomycin structure (red) with the PPARγ–rosiglitazone structure (blue). Ionomycin is in green and rosiglitazone is in yellow. (d) Superimposition of ionomycin (green) with rosiglitazone (yellow). See Jin and Li (2010) [2] for details of helix designation
Although the global structure of ionomycin-bound PPARγ is conserved to that of rosiglitazone, the binding of ionomycin triggers serial changes in the ligand-binding pocket; backbone shifts and conformational changes of the side chains induced by ionomycin contribute to this (Fig. 7). In response to the larger size of ionomycin, the backbones of helix 12 and helix 7 were both shifted outward to make extra space for ionomycin in comparison with rosiglitazone-bound PPARγ LBD (Fig. 7a, b). In addition, ionomycin also induced conformational changes of the side chains of several PPARγ pocket residues, such as F282 in helix 3, L330 in helix 5, F360 and F363 in helix 6, to accommodate the binding of ionomycin, which has a distinct chemical structure from that of rosiglitazone (Fig. 7c–f). Compared with other ligands of PPARγ, the conformational changes of backbones and side chains induced by ionomycin indicate that PPARγ has a great flexibility to adapt to the binding of diverse ligands.
Conformational changes of PPARγ LBD induced by ionomycin binding. (a, b) Overlap of the PPARγ–ionomycin structure (red) with the PPARγ–rosiglitazone (blue) structure. Ionomycin is shown in green and rosiglitazone is in yellow. The arrows indicate backbone shift changes induced by ionomycin. (c–f) Conformational changes (indicated by arrows) of the residue side chains in the PPARγ pocket involved in ionomycin binding. The electron density is shown in blue at 1σ contoured level. F360, F282 and F363 are phenylalanine at corresponding positions, L330 is leucine at position 330. See Jin and Li (2010) [2] for details of helix designation
Functional correlation of the PPARγ/ionomycin interactions
PPARγ agonists, like rosiglitazone, display a common binding mode in which the ligand forms direct contacts, such as specific hydrogen bonds with the residues on helix 12, thereby stabilising the helix 12, which is essential for a canonical active conformation of PPARγ LBD [33]. For example, rosiglitazone forms a hydrogen bond with Y473 on helix 12 through its nitrogen atom on the TZD group (Fig. 8a). However, the binding of ionomycin makes this critical Y473 residue shift outside, resulting in the loss of contacts of this residue with the ligand (Fig. 8a). In addition, ionomycin has hydrophobic groups at the corresponding position of the TZD group of rosiglitazone, making it impossible to initiate any hydrogen bond with helix 12 (Fig. 6d). All these factors may contribute to the lower degree of stabilisation of helix 12, which provides a structural support for ionomycin as a partial agonist. Accordingly, the ionomycin-mediated PPARγ transcriptional activity was substantially enhanced by the Y473F mutation while rosiglitazone-mediated activity was diminished (Fig. 8f).
Comparative analysis of the structure–activity relationship between PPARγ/ionomycin and PPARγ/rosiglitazone interactions. (a–e) Molecular determinants of the interaction between PPARγ with ionomycin and rosiglitazone in overlap. The PPARγ–ionomycin structure is shown in red while the PPARγ–rosiglitazone structure is in blue. Ionomycin is in green and rosiglitazone is in yellow. The hydrophobic interactions and hydrogen bonds are shown using lines and arrows, respectively. The potential hydrophobic interactions and hydrogen bonds, initiated by the corresponding mutations indicated in (f), are shown in dashed lines and dashed arrows, respectively. (f) Effects of mutations of key PPARγ residues on its transcriptional activity in response to ionomycin (black bars) or rosiglitazone (hatched bars) in cell-based reporter assays. Cos7 cells were cotransfected with plasmids encoding full-length PPARγ or PPARγ mutants as indicated together with a PPRE luciferase reporter. The cells were treated with 5 μmol/l ionomycin and 1 μmol/l rosiglitazone, respectively. The dotted red line indicates the activation level of wild-type (WT) PPARγ by ionomycin. Y473, I326, C285, S289, R288 and L333 are tyrosine, isoleucine, cysteine, serine, arginine and leucine at corresponding positions. Values are the means±SEM of three independent experiments
To further validate the differential roles of pocket residues in ionomycin binding and PPARγ activation, we superimposed the structure of PPARγ/ionomycin with that of PPARγ/rosiglitazone and scrutinised the PPARγ residues that contacted with different groups of ionomycin and rosiglitazone. The hydrophobic side chain of I326 makes hydrophobic interactions with the hydrophobic groups of both ionomycin and rosiglitazone (Fig. 8b). Consequently, the I326R mutation disrupting these interactions substantially reduced PPARγ transcriptional activity mediated by both ionomycin and rosiglitazone (Fig. 8f). The S289W mutation was designed to reduce the size of the PPARγ pocket, thereby preventing the binding of both ionomycin and rosiglitazone (Fig. 8c). Similar to I326R, the reduced PPARγ transcriptional activity was observed in S289W for both ionomycin and rosiglitazone (Fig. 8f). The C285A mutation was designed to increase the size of the PPARγ pocket to favour ligand binding (Fig. 8c) and L333R was designed to make the residue form hydrogen bonds with both ionomycin and rosiglitazone (Fig. 8e). Contrary to I326R and S289W, C285A and L333R substantially increased the PPARγ transcriptional activity mediated by both ionomycin and rosiglitazone (Fig. 8f). These results support a critical conserved mechanism involving the size and hydrophobic nature of the PPARγ pocket in ligand recognition.
As shown in Fig. 8d, the carboxylate oxygen from ionomycin forms indirect hydrogen bonds with R288 of PPARγ while rosiglitazone has no contacts with this residue. Mutation of R288 to hydrophobic leucine diminished PPARγ activation by ionomycin but had no effect on PPARγ activation by rosiglitazone (Fig. 8f), further highlighting the differential roles of PPARγ residues in recognising ionomycin and rosiglitazone.
Discussion
Ionomycin is an antibiotic produced by Streptomyces conglobatus, which has been characterised as a calcium ionophore [34]. In this study, we found by high-throughput screening that ionomycin is a modulating ligand for PPARγ, thereby also uncovering a novel signalling route for this antibiotic drug. The results from both biochemical AlphaScreen assay and cell-based reporter assay showed that ionomycin is a partial agonist for PPARγ due to its much lesser capability in recruiting coactivators and activating the transcriptional activity of PPARγ compared with the typical full agonist rosiglitazone. In our study, ionomycin displayed powerful anti-diabetic effects similar to TZDs; its potency correlates very well with its ability to block CDK5-mediated phosphorylation of PPARγ, supporting the critical role of PPARγ phosphorylation by CDK5 in improving insulin resistance by PPARγ ligands.
Evidence from both in vitro and in vivo studies demonstrates that ionomycin has several key features that distinguish it from the full agonist rosiglitazone. First, the scaffold of ionomycin is distinct from TZDs, with a molecular mass almost twice that of rosiglitazone, and thus represents an alternative drug design approach targeting PPARγ. Further, ionomycin takes a unique U-shape conformation within the PPARγ pocket, while SPPARMs have either one branch like MCC 555 or two branches like GW0072 (ESM Fig. 6a) [17, 35]. The specific interactions between ionomycin and the critical LBD residues of PPARγ provide critical perspectives regarding the recognition of ionomycin by PPARγ. Our structural observations indicate the loss of interactions between ionomycin and several residues on PPARγ, including one critical epitope on the activation function 2 (AF-2) helix that is used by both natural PPARγ ligands and synthetic TZDs [36, 37]; thereby, ionomycin-induced PPARγ transcriptional activity is affected. In addition, SPPARMs, such as amorfrutin 1 and GW0072, contact helix 3 and β sheets of PPARγ using their tails [35, 38], while ionomycin has no presence at the corresponding position. Instead, ionomycin has wider contacts with other regions of PPARγ including helix 7 and helix 10 (ESM Fig. 6b). Taken together, the unique characteristics of ionomycin may represent new pharmacophores that can be optimised for selectively targeting PPARγ.
More importantly, ionomycin had little adipogenic activity. A significant consequence of the transcriptional activation of PPARγ is induction of adipocyte differentiation. PPARγ agonists, including rosiglitazone, have adipogenic potency and this is the major factor leading to their adverse effects. Evidence from both 3T3-L1 cells and mouse models indicates that the adipogenic activity of ionomycin is notably weaker than that of TZDs, suggesting less adverse effects. However, more detailed clinically applied assays still need to be performed on liver toxicity and other physiological variables in vivo, such as packed cell volume for body fluid retention, to obtain a comprehensive view on the potential side effects of ionomycin. Nevertheless, our observations provide new drug design strategies for future pharmacological agents targeting PPARγ for increasing insulin sensitivity, yet without side effects such as weight gain caused by PPARγ agonism. Our structural analysis has further revealed strategies to optimise the ionomycin structure targeting PPARγ for potential therapeutic usage. For example, the chemical groups such as the carboxyl group at the ionomycin tail are essential for ion binding, which is critical for the antibiotic action of ionomycin in interfering with bacterial membrane permeability. As such, chemical groups on other PPARγ ligands and selective PPARγ modulators may be considered for replacing the corresponding ionomycin groups based on the structural analysis (ESM Fig. 6). Overall, the optimisation of the ionomycin structure to retain its ability to interact with PPARγ but reduce its ion-binding ability may yield promising therapeutic agents targeting PPARγ.
Accession number
Coordinates and structure factors for the PPARγ/ionomycin complex are available in the Protein Data Bank (www.rcsb.org/) under ID code 4FGY.
Abbreviations
- CDK5:
-
Cyclin-dependent kinase 5
- C/EBP:
-
CCAAT-enhancer binding protein
- GST:
-
Glutathione S-transferase
- GTT:
-
Glucose tolerance test
- HBC:
-
2-Hydroxypropyl-β-cyclodextrin
- mIMCD-3:
-
Mouse renal inner medullary collecting duct
- IPTG:
-
Isopropyl 1-thio-β-d-galactopyranoside
- ITT:
-
Insulin tolerance test
- LBD:
-
Ligand-binding domain
- NCoR:
-
Nuclear receptor corepressor
- NR:
-
Nuclear receptor
- PPAR:
-
Peroxisome proliferator-activated receptor
- PPRE:
-
Peroxisome proliferator hormone response element
- SCD1:
-
Stearoyl-CoA desaturase-1
- SPPARM:
-
Selective PPARγ modulator
- SRC:
-
Steroid receptor coactivator
- TZD:
-
Thiazolidinedione
References
Michalik L, Auwerx J, Berger JP et al (2006) International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol Rev 58:726–741
Jin L, Li Y (2010) Structural and functional insights into nuclear receptor signaling. Adv Drug Deliv Rev 62:1218–1226
Tontonoz P, Spiegelman BM (2008) Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem 77:289–312
Kuda O, Jelenik T, Jilkova Z et al (2009) n-3 fatty acids and rosiglitazone improve insulin sensitivity through additive stimulatory effects on muscle glycogen synthesis in mice fed a high-fat diet. Diabetologia 52:941–951
Lennon R, Welsh GI, Singh A et al (2009) Rosiglitazone enhances glucose uptake in glomerular podocytes using the glucose transporter GLUT1. Diabetologia 52:1944–1952
Serrano-Marco L, Barroso E, El Kochairi I et al (2012) The peroxisome proliferator-activated receptor (PPAR) β/δ agonist GW501516 inhibits IL-6-induced signal transducer and activator of transcription 3 (STAT3) activation and insulin resistance in human liver cells. Diabetologia 55:743–751
Lehrke M, Lazar MA (2005) The many faces of PPARgamma. Cell 123:993–999
Wang YX (2010) PPARs: diverse regulators in energy metabolism and metabolic diseases. Cell Res 20:124–137
Weismann D, Erion DM, Ignatova-Todorava I et al (2011) Knockdown of the gene encoding Drosophila tribbles homologue 3 (Trib3) improves insulin sensitivity through peroxisome proliferator-activated receptor-γ (PPAR-γ) activation in a rat model of insulin resistance. Diabetologia 54:935–944
Yki-Jarvinen H (2004) Thiazolidinediones. N Engl J Med 351:1106–1118
Nissen SE, Wolski K (2007) Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. N Engl J Med 356:2457–2471
Zanchi A, Maillard M, Jornayvaz FR et al (2010) Effects of the peroxisome proliferator-activated receptor (PPAR)-gamma agonist pioglitazone on renal and hormonal responses to salt in diabetic and hypertensive individuals. Diabetologia 53:1568–1575
Evans RM, Barish GD, Wang YX (2004) PPARs and the complex journey to obesity. Nat Med 10:355–361
Feige JN, Gelman L, Michalik L, Desvergne B, Wahli W (2006) From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Prog Lipid Res 45:120–159
Moras D, Gronemeyer H (1998) The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol 10:384–391
Allen T, Zhang F, Moodie SA et al (2006) Halofenate is a selective peroxisome proliferator-activated receptor gamma modulator with antidiabetic activity. Diabetes 55:2523–2533
Zhang F, Lavan BE, Gregoire FM (2007) Selective modulators of PPAR-γ activity: molecular aspects related to obesity and side-effects. PPAR Res 2007:32696
Higgins LS, Depaoli AM (2010) Selective peroxisome proliferator-activated receptor gamma (PPARgamma) modulation as a strategy for safer therapeutic PPARgamma activation. Am J Clin Nutr 91:267S–272S
Choi JH, Banks AS, Estall JL et al (2010) Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature 466:451–456
Houtkooper RH, Auwerx J (2010) Obesity: new life for antidiabetic drugs. Nature 466:443–444
Li P, Fan W, Xu J et al (2011) Adipocyte NCoR knockout decreases PPARγ phosphorylation and enhances PPARγ activity and insulin sensitivity. Cell 147:815–826
Otwinowski Z, Minor W (1997) Processing of x-ray diffraction data collected in oscillation mode. Method Enzymol 276:307–326
Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132
Li Y, Suino K, Daugherty J, Xu HE (2005) Structural and biochemical mechanisms for the specificity of hormone binding and coactivator assembly by mineralocorticoid receptor. Mol Cell 19:367–380
Li Y, Choi M, Cavey G et al (2005) Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Mol Cell 17:491–502
Jin L, Lin S, Rong H et al (2011) Structural basis for iloprost as a dual peroxisome proliferator-activated receptor alpha/delta agonist. J Biol Chem 286:31473–31479
Lin S, Han Y, Shi Y et al (2012) Revealing a steroid receptor ligand as a unique PPARgamma agonist. Cell Res 22:746–756
Leesnitzer LM, Parks DJ, Bledsoe RK et al (2002) Functional consequences of cysteine modification in the ligand binding sites of peroxisome proliferator activated receptors by GW9662. Biochemistry 41:6640–6650
Tontonoz P, Hu E, Spiegelman BM (1994) Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 79:1147–1156
Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM (1994) mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev 8:1224–1234
Gampe RT Jr, Montana VG, Lambert MH et al (2000) Asymmetry in the PPARgamma/RXRalpha crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell 5:545–555
Chandra V, Huang P, Hamuro Y et al (2008) Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature 456:350–356
Nolte RT, Wisely GB, Westin S et al (1998) Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395:137–143
Liu C, Hermann TE (1978) Characterization of ionomycin as a calcium ionophore. J Biol Chem 253:5892–5894
Oberfield JL, Collins JL, Holmes CP et al (1999) A peroxisome proliferator-activated receptor gamma ligand inhibits adipocyte differentiation. Proc Natl Acad Sci USA 96:6102–6106
Itoh T, Fairall L, Amin K et al (2008) Structural basis for the activation of PPARgamma by oxidized fatty acids. Nat Struct Mol Biol 15:924–931
Li Y, Zhang J, Schopfer FJ et al (2008) Molecular recognition of nitrated fatty acids by PPAR gamma. Nat Struct Mol Biol 15:865–867
Weidner C, de Groot JC, Prasad A et al (2012) Amorfrutins are potent antidiabetic dietary natural products. Proc Natl Acad Sci USA 109:7257–7262
Acknowledgements
We thank the staff at BL17U of the Shanghai Synchrotron Radiation Source for assistance in data collection.
Funding
This work was supported by grants from the National Basic Research Program of China (973 Programme: 2012CB910104 to Y. Li), the National Natural Science Foundation of China (31070646 and 31270776 to Y. Li), the Fundamental Research Funds for the Central Universities (2010121083 to L. Jin, 2011121029 to Y. Li) and the Programme of Introducing Talents of Discipline to Universities (B12001).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
WZ and XF researched the data and drafted the article. LJ and YL designed and researched the data, and wrote the article. LQ, ZP, RW, SL and DH researched data and revised the article. All authors approved the final version to be published.
Author information
Authors and Affiliations
Corresponding authors
Additional information
W. Zheng and X. Feng contributed equally to this study.
Electronic supplementary materials
Below is the link to the electronic supplementary material.
ESM Table 1
(PDF 59 kb)
ESM Table 2
(PDF 50 kb)
ESM Table 3
(PDF 85 kb)
ESM Fig. 1
(PDF 80 kb)
ESM Fig. 2
(PDF 28 kb)
ESM Fig. 3
(PDF 74 kb)
ESM Fig. 4
(PDF 244 kb)
ESM Fig. 5
(PDF 108 kb)
ESM Fig. 6
(PDF 333 kb)
Rights and permissions
About this article
Cite this article
Zheng, W., Feng, X., Qiu, L. et al. Identification of the antibiotic ionomycin as an unexpected peroxisome proliferator-activated receptor γ (PPARγ) ligand with a unique binding mode and effective glucose-lowering activity in a mouse model of diabetes. Diabetologia 56, 401–411 (2013). https://doi.org/10.1007/s00125-012-2777-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-012-2777-9