Abstract
The highly specific induction of RNA interference-mediated gene knockdown, based on the direct application of small interfering RNAs (siRNAs), opens novel avenues towards innovative therapies. Two decades after the discovery of the RNA interference mechanism, the first siRNA drugs received approval for clinical use by the US Food and Drug Administration and the European Medicines Agency between 2018 and 2022. These are mainly based on an siRNA conjugation with a targeting moiety for liver hepatocytes, N-acetylgalactosamine, and cover the treatment of acute hepatic porphyria, transthyretin-mediated amyloidosis, hypercholesterolemia, and primary hyperoxaluria type 1. Still, the development of siRNA therapeutics faces several challenges and issues, including the definition of optimal siRNAs in terms of target, sequence, and chemical modifications, siRNA delivery to its intended site of action, and the absence of unspecific off-target effects. Further siRNA drugs are in clinical studies, based on different delivery systems and covering a wide range of different pathologies including metabolic diseases, hematology, infectious diseases, oncology, ocular diseases, and others. This article reviews the knowledge on siRNA design and chemical modification, as well as issues related to siRNA delivery that may be addressed using different delivery systems. Details on the mode of action and clinical status of the various siRNA therapeutics are provided, before giving an outlook on issues regarding the future of siRNA drugs and on their potential as one emerging standard modality in pharmacotherapy. Notably, this may also cover otherwise un-druggable diseases, the definition of non-coding RNAs as targets, and novel concepts of personalized and combination treatment regimens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Gene knockdown via RNA interference can be induced by small interfering RNAs and opens novel avenues towards innovative therapies in many diseases. |
Despite their very attractive mechanism of action, the development of small interfering RNA therapeutics faces several challenges and issues. |
Two decades after the discovery of the RNA interference mechanism, the first small interfering RNA drugs have received approval for clinical use and several other small interfering RNAs are in late-stage clinical studies. |
1 The History: RNAi/siRNA Discovered, Not Invented
The concept of RNA interference (RNAi) was discovered in 1998 as a naturally occurring defense mechanism against the invasion of foreign nucleic acids and the control of gene expression [1]. Soon thereafter, small interfering RNAs (siRNAs) were identified as mediators of RNAi in mammalian cells [2]. Moreover, it was found that the delivery of siRNAs is necessary and sufficient for inducing RNAi-mediated gene knockdown, as the other components of the RNAi machinery are provided by the cell. It thus quickly became obvious that RNAi based on the direct application of siRNAs opens novel avenues towards innovative therapies. The availability of powerful siRNA in silico prediction tools and comprehensive human genome data allows for the straightforward definition of siRNAs against a given target gene. This soon led to the first attempts to explore siRNAs as therapeutics. These, however, were associated with several setbacks, mainly owing to issues regarding siRNA delivery, stability, specificity, and unwanted side effects.
2 The Beauty: The RNAi Mechanism Different from Other Drug Actions
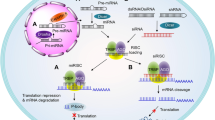
The details of the RNAi mechanism have been extensively studied; the reader is referred to many excellent reviews on this topic and to Fig. 1 for illustration. Briefly, the endoribonuclease Dicer processes longer double-stranded RNA or short-hairpin RNA into mature siRNA [3]. This siRNA, which can also be delivered directly to the cell as a chemically synthesized molecule, is introduced in the RNA-induced silencing complex (RISC) that comprises several distinct proteins including Argonaute-2 (Ago-2) and Dicer [4]. After siRNA activation by removing its ‘sense’ or ‘passenger’ strand, the remaining ‘antisense’ or ‘guide’ strand directs RISC towards binding to the target messenger RNA (mRNA), where Ago-2 in RISC mediates cleavage [5]. In contrast to antisense technologies, RNAi relies on a catalytic mechanism, as, after target mRNA cleavage, siRNA-loaded RISC is able to dissociate and bind to another mRNA molecule. In consequence, very low siRNA concentrations in the picomolar range are able to induce efficient gene knockdown and intracellular amounts of less than 2000 siRNAs per cell have been determined as sufficient [6]. The mechanism should also be distinguished from microRNAs (miRNAs), which bind with only partial complementarity while siRNA action relies on 100% complementarity.
Schematic of the mechanism of synthetic small interfering RNA (siRNA)-mediated knockdown. Synthetic mature siRNA can be effectively delivered to the cell by nanocarriers, as an siRNA conjugate or through other mechanisms (see text for details). The double-stranded siRNA comprises the ‘sense’ or ‘passenger strand’ (red) and the ‘antisense’ or ‘guide strand’ (blue). After cell entry, for example, via endocytosis and escape from the endosome, the siRNA is introduced into the RNA-induced silencing complex (RISC), which comprises several distinct proteins including Argonaute-2 (Ago-2), Dicer, and the transactivation response element RNA-binding protein (TRBP). Upon siRNA activation by removing its ‘sense’ or ‘passenger’ strand, the remaining ‘antisense’ or ‘guide’ strand directs RISC towards sequence-specific binding to the target messenger RNA (mRNA). The siRNA action relies on 100% complementarity to the target sequence, and by bringing RISC into close proximity to its target mRNA, it initiates Ago-2-mediated mRNA cleavage (black scissors). Because of this cleavage and the subsequent presence of unprotected ends, the mRNA is rapidly degraded by intracellular RNAses, leading to the efficient prevention of protein synthesis. After mRNA cleavage, the siRNA-loaded RISC can dissociate and bind to another mRNA target molecule, thus acting in a catalytical manner
The fact that siRNAs thus act post-transcriptionally on the mRNA level rather than post-translationally on proteins allows for inhibiting targets for which no inhibitors are available or possible to develop and which are therefore considered as ‘undruggable genes’ [7]. This profoundly extends the spectrum of potential targets beyond the small fraction of druggable proteins [8].
3 The Challenge: Developing the Optimal siRNA
The optimal siRNA should have the following characteristics: no activation of the innate immune system, efficient and specific cleavage of its target, no off-target (i.e., effects on non-target genes) or other toxic effects, and long half-life/slow degradation in the body circulation and inside the target cells. From a therapeutic point of view, the first important aspect is the selection of the right target to be silenced for treating a particular disease. So far, mainly protein-coding mRNAs have been therapeutically addressed by siRNAs [9]. Since the Encyclopedia of DNA Elements (ENCODE) project in 2012 [10], it is known that a major part of the human genome encodes for non-protein coding RNAs. Among those, long noncoding RNAs are of particular interest from a clinical viewpoint, comprising many novel putative therapeutic targets [10], especially for the prospective treatment of cancer [11, 12]. Likewise, RNAi is a promising therapeutic approach to fight infections caused by pathogenic viruses featuring single-stranded RNA genomes and subgenomic RNA transcripts (e.g., hepatitis C virus, respiratory syncytial virus, influenza, coronaviruses) [13,14,15,16]. The worldwide rampant severe acute respiratory syndrome coronavirus 2 pandemic has especially catalyzed the progress in promising antiviral siRNA therapy development [17,18,19,20,21].
The first protocol for designing an effective siRNA was published in 2001 [22]. Therein, Elbashir et al. recommended the use of siRNAs with 21 nucleotides in length, with a G/C content of ~ 50% and 3′-overhangs of two nucleotides. Since then, extensive research has led to many improvements in siRNA design, which is currently based on sophisticated algorithms (see below and Fig. 2).
Schematic of important features and parameters in the small interfering RNA (siRNA) design. General important features for an optimal siRNA design (upper boxes) and the identification of optimal messenger RNA (mRNA) target positions (lower box) are described. The scheme of the double-stranded siRNA (center; red, ‘sense’ or ‘passenger’ strand; blue, ‘antisense’ or ‘guide’ strand) shows the characteristic deoxyribonucleic acid overhangs at the 3′-end (e.g., dT; black circles), but does not include any chemical modifications (for details, see below). dsRNA double-stranded RNA, miRNA microRNA, ORF open reading frame, UTR untranslated region
The length of siRNAs now used ranges from 19 to 29 nucleotides [23, 24]. Short siRNAs may tend towards more unspecific binding, but in the range of 19–25 nucleotides siRNAs show similar efficiency in gene silencing [25]. The use of short siRNA is preferred because longer siRNAs can provoke an inflammatory antiviral immune response [26]. Currently, however, the introduction of chemical nucleotide modifications can effectively prevent this unwanted side effect (reviewed in [27]). Synthetic RNA entry can erroneously be recognized as viral RNAs by endosomal and intracellular host pattern recognition receptors of the innate immune system (e.g., Toll-like receptors [TLR-3, TLR-8, and TLR-9], retinoic acid inducible gene I receptor, and double-stranded RNA-activated protein kinase) [26, 28, 29]. Type I interferons (interferon-α and interferon-β) and inflammatory cytokines are produced as antiviral immune response [30]. The activated protein kinase leads to the inhibition of mRNA translation and cell death by phosphorylation of eukaryotic translation initiation factor 2-alpha. To prevent activation of the immune system, specific sequence patterns or high “U” content (recognized by TLRs) should be avoided. Instead, shorter (< 24 bp) or chemically modified siRNA can be used (preventing activated protein kinase) and 3′-overhangs allow evasion from retinoic acid inducible gene I receptor immune sensing.
To design the most effective siRNAs, essential parameters and position-specific nucleotide preferences have been uncovered in numerous biological and bioinformatics studies. Several protocols have been published, and online available algorithms or proprietary software programs assist in the design of new siRNAs [25, 31,32,33,34,35,36,37].
As a basic parameter, the GC content of an siRNA is addressed by algorithms and its range should be between ~30 and 60% [33, 34]. Too low GC content can lead to weak or unspecific binding, whereas too high GC content may impede unwinding by helicase and incorporation in the RISC complex [25]. Between nucleotides 9 and 14, however, low GC content is important to increase RISC function during mRNA cleavage [38]. Sequences that could lead to secondary structures in the sense or antisense stand must be avoided (e.g., internal repeats, palindromes, CCC or GGG sequences) [32, 34, 39]. A proper duplex formation is essential for functional siRNA. Additionally, sequences that contain single nucleotide polymorphisms, miRNA seed matches, and known toxic motifs must be avoided [40,41,42]. An asymmetrical nucleotide content in the duplex and weak base pairing at the 5′-end of the antisense strand is very important to ensure preferential loading of the antisense strand into the RISC complex (‘strand bias’) [31, 34]. Therefore, more A/U at the 5′-end of the antisense strand and more G/C at the 5′-end of the sense strand reduce the risk of sense strand incorporation into RISC and the associated off-target effects. The strongest correlation with siRNA efficiency was found for the presence of U at position 10 of the sense strand [34]. This seems to be because RISC cleaves its target mRNA between nucleotide 10 and 11, with a preference for cutting 3′ of U rather than other bases [25, 43]. Further rules include the presence of A at positions 4 and 19 of the sense strand, the absence of G at position 13 and of G/C at position 19 of the sense strand, and the presence of A at position 6 of antisense strand. A dTdT nucleotide overhang at the 3′-end enhances resistance of siRNA duplexes to degradation by RNAse [31, 34, 35].
Despite a long controversial debate regarding the influence of the target mRNA structure on siRNA efficiency, many studies uncovered a direct correlation of the local target secondary structure and target site accessibility on RNAi efficiency [44,45,46,47,48]. In this context, the 5′-untranslated region (and 3′-untranslated region) of mRNA as well as sequences close to the start codon are not recommended as siRNA targets, as the binding of regulatory proteins in this area may impede RISC binding and thus the silencing effect. Rather, selecting regions in the open reading frame about 50–100 nucleotides downstream of the start codon is recommended. Furthermore, siRNAs closer to the start codon seem to be more efficient than those further downstream [25].
If different mRNA transcript variants exist, generated by alternative splicing during transcription, use of alternative promoters or alternative polyadenylation sites, sequences that are not present in all relevant variants must be excluded. The following transcript databases and browsers assist in these analyses: NCBI (https://www.ncbi.nlm.nih.gov) [49], Ensembl (https://www.ensembl.org) [50], and UCSC Genome (https://genome.ucsc.edu) [51]. Therapeutic efficacies will highly rely on the selection of a functionally relevant target gene. In oncology, this may be based on high levels of (over-)expression or the identification of critical driver mutations. For viral RNA targets, it is crucial to identify suitable regions with nucleotide sequences highly conserved between variants or strains, which are not subject to high selection pressure and contain functionally essential components of the virus [21].
4 The Chemistry: siRNA Modifications
To enhance siRNA functionality and improve other siRNA properties, various nucleotide modifications have been developed and implemented in siRNA design. As basic building blocks of RNA, natural nucleotides usually consist of a ribose sugar, with a 1′-nucleobase group and a 3′-phosphate group. Based on the structure of an RNA nucleotide, it can be distinguished between phosphonate modifications (Fig. 3A), base modifications (Fig. 3B), and ribose modifications (Fig. 3C). Chemical modifications of siRNA nucleotides and the use of analogs provide solutions to many of the challenges in the development of siRNA therapeutics. Although completely unmodified or slightly modified siRNAs are able to mediate gene silencing in vivo (especially in tissues where local application is possible), extensive modifications can (i) efficiently suppress immunostimulatory siRNA-driven activation of innate immune response, (ii) improve chemical stability and efficacy, and (iii) decrease off-target-induced toxicity [52, 53]. Based on the natural structure of nucleotides, chemical modifications can be implemented in the ribose moiety, the phosphate backbone, and the bases (Fig. 3).

Adapted from Hu et al. [27]
Overview of chemical oligonucleotide modifications relevant for small interfering RNA. Based on the structure of an RNA nucleotide, it can be distinguished between A phosphonate modifications, B base modifications, and C ribose modifications.
Ribose modifications (Fig. 3C) at the 2′-position are the most common and widely used for protecting siRNA from a ribonuclease attack [27]. As the 2′-OH group is essential for enzymatic hydrolysis, but not required for RNA activity, it can be replaced for example by 2′-O-methyl (2′-O-Me; a natural ribose sugar). 2′-O-methyl is the most frequently used siRNA modification [54], which significantly increases not only the siRNA stability and half-life, but also siRNA affinity to the target mRNA [55], while simultaneously reducing immunogenicity. A series of other analogs have been introduced as well, such as 2′-O-methoxyethyl as another very useful and popular analog, mediating higher siRNA binding affinity to RNA and resistance towards a nuclease attack [53]. The substitution of the 2′-OH group by highly electronegative fluorine like 2′-deoxy-2′-fluoro (2′-F) or 2′-arabino-fluoro is very efficient as well [56]. The 2′-F modification is widely employed in clinically and preclinically used siRNAs and confers a C3-endo conformation beneficial for binding activity. 2′-O-benzyl and 2′-O-methyl-4-pyridine confer increased activity when placed at positions 8 and 15 of the siRNA guide strand [57]. Other modifications at the 2′-C, 4′-C, or the whole sugar ring include unlocked nucleic acids, locked nucleic acids (LNAs), glycol nucleic acid, constrained 5-methyluracil nucleoside (S)-cEt-BNAs, tricyclo-DNA, and phosphorodiamidate morpholino oligomers. Both unlocked nucleic acids and glycol nucleic acid act as thermally destabilizing nucleotides that minimize off-target effects by blocking the entry of the passenger strand and promoting RISC loading of the guide strand, thus introducing chemical asymmetry into duplex siRNAs. The last ribose modification to be mentioned here is LNAs, i.e., a bicyclic furanose unit locked in an RNA mimicking sugar conformation. Locked nucleic acids significantly increase the affinity of base pairing by “locking” the ribose into its preferred C3′-endo conformation [58].
Many phosphate modifications are used as well (Fig. 3A), for example, phosphorothioate (PS as Rp or Sp isomer, or at the 5′-end), phosphorodithioate, methylphosphonate, methoxypropylphosphonate, 5′-(E)-vinylphosphonate, and peptide nucleic acid. The PS linkage is obtained by a sulfur atom replacing one non-bridging oxygen of the phosphodiester and was initially introduced in antisense oligonucleotide (ASO) modification [59, 60]. These modifications confer resistance to nucleases, enhance hydrophobic protein-binding properties (e.g., to blood serum proteins), extend the half-life of modified oligonucleotides in the circulation and seem to be beneficial for their cell entry, while barely affecting in vivo biodistribution profiles (predominant accumulation in excretion organs) [61, 62]. In contrast, substantial PS modification was shown to reduce to some extent the oligonucleotide binding affinity to its target sequence [53], and excessive protein binding was found to be associated with in vivo toxicity [63, 64]. Still, PS modification is very important and necessary for both, ASO and siRNA. In fact, the clinically approved siRNA therapeutics givosiran, lumasiran, and inclisiran (all from Alnylam Phamaceuticals) contain PS modifications. More specifically, Alnylam introduced two PS linkages at the first two nucleotides at the 5′-ends of the sense strand and of the antisense strand, respectively, and two additional PS linkages at the first two nucleotides at the 3′-end of the antisense strand of the siRNA [27]. The PS2 modification increases the affinity between RISC and siRNA. For further enhancement of siRNA stability, analogs with a conformation similar to natural phosphates, but resistant to dephosphorylation have been identified. Among them, the 5′-(E)-VP modification is the most potent and metabolically stable modification. Additionally, siRNA carrying an (E)-VP at the 5′-end of the guide strand augments gene silencing by enhanced binding to human Ago-2 [65,66,67].
Base modifications or replacements (Fig. 3B) are not widely used for siRNA and ASO modifications, and up to now rather a subject of research. However, these modifications may provide prospective benefits and opportunities to improve drug development. Indeed, innate immune recognition of ASO can be reduced and resistance to nucleases can be improved when using the base analogs 2-thiouridine, N6-methyladenosine, and 5-methylcytidine, or other base analogs of uridine and cytidine residues. Because the safety of these non-natural residues in metabolism remains unclear, the use of naturally occurring base structures, for example, 5mC or 6mA, is currently preferred [27]. The use of N-ethylpiperidine triazole-modified adenosine analogs for siRNA modification is able to disrupt nucleotide/TLR8 interactions and therefore decreases the immunogenicity of siRNA [68, 69]. A practical strategy to reduce the passenger strand-mediated off-target effects might be the 5-nitroindole modification at position 15 of the siRNA passenger strand. Notably, siRNAs containing 5-fluoro-2′-deoxyuridine moieties may provide an additional strategy for siRNA-based cancer therapy, by quickly releasing 5-fluoro-2′-deoxyuridine after cell entry with a subsequent induction of a variety of DNA-damage repair and apoptosis pathways that trigger cell death [70].
The following siRNA modifications and combinations thereof are frequently used in siRNA therapeutics approved for clinical use or currently in clinical studies (Fig. 4). These include 2′-O-Me (e.g., QPI-1002 (I5NP) [71] and QPI-1007 [72]; Quark Pharmaceuticals), 2′-O-Me and 2′-F (e.g., Partisiran [Onpattro®] [73]; Alnylam Pharmaceuticals), 2′-O-Me, 2′-F and PS (e.g., Givosiran [Givlaari®] [74, 75], Inclisiran [Leqvio®] [76], Lumasiran [Oxlumo®] [77]; Alnylam Pharmaceuticals), 2′-O-Me, 2′-F, PS, and inverted Base (e.g., ARO-AAT [78] and ARO-APOC3 [79]; Arrowhead Pharmaceuticals), and 2′-O-Me, 2′-F, PS, and glycol nucleic acid (e.g., ALN-HBV02 [VIR-2218] [80]). In initial therapeutic siRNA developments, unmodified or partially modified (e.g., Partisiran [Onpattro®]) siRNAs were typically used. As a result of scientific progress, however, most siRNAs are currently fully modified (e.g., givosiran [Givlaari®], inclisiran [Leqvio®], lumasiran [Oxlumo®], vutrisiran [Amvuttra®]). In this context, various modification patterns have been commercially developed, such as standard template chemistry, enhanced stabilization chemistry (ESC), advanced ESC and ESC plus (all established by Alnylam Pharmaceuticals), or designs established by Arrowhead Pharmaceuticals and Dicerna Pharmaceuticals, respectively (reviewed in [27]).

Adapted from Hu et al. [27]
Chemical modification patterns of small interfering RNAs approved for clinical use: patisiran, givosiran, inclisiran, lumasiran, and vutrisiran. Modifications are indicated by colored circles, and phosphorothioate modifications are highlighted as well; see the lower panel for an explanation of the symbols.
During the siRNA design, sequences of both strands (antisense and sense) need to be checked for sequence specificity via a BLAST analysis with a reference sequence database (Refseq-RNA database). Sequences that have perfect or near-perfect complementarity to unintended targets should be discarded to reduce the risk of off-target effects. A query coverage with other genes of less than 78% and a match of ≤ 15 out of 19 nucleotides is considered tolerable [25, 81]. To exclude the probability of unpredictable off-target effects, siRNA knockdown studies analyzed by whole transcriptome sequencing can be performed. The effectiveness of various siRNA candidates to silence the specific target gene of interest must be experimentally verified. Although these functional gene knockdown studies can be facilitated by using reporter gene assays, it is strongly recommended to include appropriate target cells that endogenously express the target gene [82]. Comparable to other pharmaceutical drugs, market authorization requires preclinical safety and efficacy studies in vitro and in vivo, animal models, biodistribution patterns, and clinical studies. For example, regulatory guidelines and preclinical tools related to RNA biodistribution have been reviewed recently [83].
5 One Issue: Therapeutic Delivery of siRNA
RNA molecules must be considered as instable, and prone to rapid enzymatic and non-enzymatic degradation. Moreover, they are sufficiently small (7–8 nm in length and 2–3 nm in diameter) to be subject to renal clearance while being still too large (~ 13 kD) for membrane passage and cellular uptake. The half-life of chemically unmodified naked siRNAs in the bloodstream is only ~ 5 minutes [84] and only a few cell types such as neurons or retinal ganglion cells are capable of taking up naked siRNAs. Thus, the development of delivery strategies has been a major bottleneck on the way towards using siRNAs in vivo and in particular towards their translation into the clinics. It still represents a major limitation to their therapeutic use in many pathologies. Thus, initial clinical studies of siRNA therapeutics only a very few years after their discovery focused on local treatment, for example, intravitreal injection (see [85] for review). Upon systemic application, the liver as a target organ is comparatively easy to reach and at the forefront of the development. In general, it can be distinguished between covalent coupling of targeting moieties directly to the siRNAs and delivery systems based on various nanoparticles. The latter include a wide selection of inorganic, lipid, polymeric, or other carrier systems for siRNA adsorption or encapsulation, mediating siRNA protection, cellular uptake, and intracellular processing to the correct subcellular site of action. As this is not the major topic of this article, the reader is referred to many comprehensive reviews on siRNA delivery, for example [86,87,88,89,90,91].
So far, clinical approval of siRNA drugs has been limited to siRNA conjugates with GalNAc for delivery into hepatocytes, based on high affinity binding to the asialoglycoprotein receptor (ASGPR), which is highly expressed on the surface of hepatocytes, while largely absent on other cells [92,93,94,95], and lipid-based nanoparticles (LNPs), in particular ‘stable nucleic acid lipid particles’ as second-generation LNPs [96]. Based on liposomes for the formulation of cytostatic drugs such as doxorubicine (Doxil®), daunorubicine (DaunoXome®), or vincristine (Marqibo®) that have been approved already (see e.g., [87] for review), further adaptation with regard to lipid composition and overall LNP design has led to systems adapted to siRNA delivery. In particular, second-generation systems with pH-dependent ionization properties have addressed problems related to a permanent positive nanoparticle surface charge, including clearance by the reticuloendothelial system and toxicity. These systems are essentially uncharged at a physiological pH, but acquire a positive charge at a low pH, for example, in the endosomal/lysosomal system. They are capable of interacting with apolipoprotein E3, a transporter of lipids to hepatocytes, and thus show a high efficacy for hepatocyte delivery with lower side effects compared with lipids with a permanent positive charge [97]. As a result, three lipid-based siRNA delivery systems are available from Alnylam and Arbutus, DLin-DMA, DLin-MC3-DMA, and L319. From a large screening of thousands of lipidoids obtained by a strategy based on combinatorial chemistry, other LNPs were identified as efficient [27, 98]. These and other studies have established important properties of LNPs for siRNA delivery to the liver, including the pKa of the lipids, the polyethylenglycol content and molecular weight, and the hydrophobic tails [99,100,101]. Beyond lipids, other companies have focused on other delivery systems for clinical translation (see below and Table 1).
For hepatocyte targeting, the already mentioned GalNAc is considered as superior over liposomal systems with regard to efficacy and safety. Trivalent or tetravalent GalNAc moieties are coupled to the 5′-end or 3′-end of siRNA sense strands via a linker, with triantennary GalNAc showing the highest affinity for ASGPR [102]. To avoid rapid renal clearance and achieve a better delivery performance, subcutaneous administration of GalNAc siRNAs has been found advantageous over an intravenous application. Cellular uptake is mediated by ASGPR-mediated endocytosis. Beyond Alnylam, other companies such as Arrowhead (TRiM; Targeted RNAi Molecule) and Dicerna have established GalNAc-based conjugate platforms as well, or GalNAc analogs as ASGPR ligands (see [27] and references therein). Other targeting moieties including RGD motifs, i.e., ligands to integrins αvβ3 and αvβ5 for delivery into tumor cells, or αvβ6 for reaching lung epithelial cells, are explored as well (see below, Sect. 8).
6 The Present Situation: First siRNA Drugs Approved for Clinical Use
Twenty years after the discovery of the RNAi mechanism, the first siRNA drugs have received approval for clinical use (Table 2).
6.1 Patisiran
Patisiran (Onpattro®; Alnylam) was the first siRNA drug approved in 2018 for clinical use [103,104,105]. It is directed against the transthyretin (TTR) gene, for treating hereditary variant transthyretin amyloidosis. The TTR protein is a transporter protein for thyroid hormone, thyroxine, and retinol. It is produced in the liver and leads, when mutated, to misfolding of TTR fibrils and their deposition and accumulation in various organs and tissues. Hereditary variant transthyretin amyloidosis is a rare, progressively fatal disease. Predominant symptoms are cardiomyopathy and polyneuropathy, mainly characterized by sensory, motor, and autonomic dysfunction. Treatment options are limited to liver transplantation, where applicable, and TTR tetramer-stabilizing drugs, tafamidis and diflunisal. The chemically modified siRNA is formulated in a lipid nanoparticle, comprising the cationic ionizable lipid DLin-MC3-DMA, cholesterol, the polar phospholipid DSPC, and PEG-modified lipids (PEG2000-C-DMG) [106]. After their mixing under acidic conditions, small pH-sensitive liposomes are formed that fuse into larger lipid nanoparticles upon pH neutralization. Upon an intravenous injection of patisiran, the PEG-modified lipids are replaced by serum proteins, in particular apolipoprotein, which interacts with the cholesterol component of the lipid nanoparticle and mediates targeted delivery to hepatocytes. There, the internalized lipid nanoparticles reach the endosome where, at a lower pH, DLin-MC3-DMA becomes cationic and eventually leads to endosome disruption.
Patisiran is to be administered via an intravenous injection every 3 weeks. With the recommended dose of 0.3 mg/kg, steady state is reached within 24 weeks. In order to prevent infusion-related reactions, patients are pre-medicated intravenously with an antihistamine and a corticosteroid as well as oral acetaminophen [107]. After phase I and phase II trials, approval was based on the favorable results from the large global APOLLO phase III trial [73, 108]. Based on a rapid and robust 81% decrease of serum TTR levels over the whole 18-month study period, a statistically significant improvement in polyneuropathy as determined by the modified Neuropathy Impairment Score as well as all secondary endpoints (motor strength, body mass index, level of disability, mobility, several scaring systems for measuring quality of life) was observed. It was also found to be well tolerated [107]. In a global Open-Label Extension (OLE) study, the long-term safety of patisiran is currently evaluated in adults with hereditary transthyretin-mediated amyloidosis and symptomatic polyneuropathy, who participated in a previous study. A most recent publication presented the 12-month efficacy and safety results of a prospective study of patisiran in patients with hereditary variant transthyretin amyloidosis who had polyneuropathy progression post-liver transplantation, demonstrating that the drug reduced serum TTR, was well tolerated, and improved or stabilized key disease impairment measures [109]. Phase IV observational studies (ConTTRibute) in patients with or without mutations and a pregnancy surveillance program are ongoing as well.
6.2 Givosiran
Givosiran (Givlaari®; Alnylam) was the second siRNA therapeutic to reach the market, receiving US Food and Drug Administration (FDA) approval in 2019 [110, 111]. It is directed against δ-aminolevulinic acid synthase 1 (ALAS1), which plays a critical role in acute hepatic porphyria (AHP). Acute hepatic porphyria is a rare genetic disorder based on mutations in genes involved in heme biosynthesis [112]. More specifically, the upregulation of hepatic ALAS1 leads to the accumulation of heme intermediates, δ-aminolevulinic acid, and porphilinogen, which cause neurotoxicity. This can result in chronic symptoms as well as debilitating and potentially life-threatening acute porphyria attacks (abdominal pain, nausea, vomiting, constipation, weakness, and psychiatric symptoms). Furthermore, AHP can contribute to comorbidities such as chronic neuropathy, liver disease, systemic arterial hypertension, or chronic kidney disease. Treatment options for AHP are limited and include the elimination of precipitating factors, carbohydrate loading (ALAS1 is upregulated during fasting), symptomatic supportive therapy, and hemin administration. Givosiran leads to ALAS1 knockdown in hepatocytes and thus to a reduction in circulating δ-AKA and porphilinogen levels.
Givosiran is based on ESC and GalNAc technology developed by Alnylam. Preclinical studies performed in mice, rats, and cynomolgus monkeys showed a rapid, robust, and sustained reduction in ALAS1 [113, 114], and provided the basis for clinical studies (Table 2 [74, 115]; see [116] for details). Upon subcutaneous injection, givosiran is rapidly absorbed, shows high (>90%) plasma protein binding, rapid liver uptake, and sustained knockdown activity over the dosing regimen of once per month. In the phase III ENVISION trial in patients with acute intermittent porphyria, it was found to be generally well tolerated, with an acceptable safety profile. Givosiran led to a significantly lower rate of porphyria attacks and better results for multiple other disease manifestations as compared with the placebo group. The increased efficacy, however, was accompanied by a higher frequency of hepatic and renal adverse events [75]. Based on these results from the ENVISION trial, Givosiran has been approved in the USA, the European Union, and in other countries for the treatment of AHP [110, 111]. It is administered by a subcutaneous injection once per month at a recommended dosage of 2.5 mg/kg. Additional data on long-term efficacy and safety are still needed. In this regard, results from the OLE study after completion of the ENVISION study will be of interest. The 24-month interim analysis demonstrated long-term givosiran administration to result in a sustained δ-aminolevulinic acid and porphobilinogen reduction. Long-term givosiran showed an acceptable safety profile and significant benefits in patients with AHP with recurrent attacks, based on the reduction in attack frequency, hemin use, and severity of daily worst pain while improving quality of life [117].
6.3 Inclisiran
Inclisiran (Leqvio®; Novartis) is another GalNAc-siRNA conjugate, developed for the treatment of homozygous familial hypercholesterolemia and elevated low-density lipoprotein cholesterol (LDL-C). By treating hypercholesterolemia as a very prevalent indication, its impact may well exceed those of other siRNA therapeutics, especially considering the increasing need for novel treatment options in this disease beyond statins. It targets PCSK9 (pro-protein convertase subtilisin/kexin type 9), a key enzyme in the LDL-C metabolic pathway. More specifically, knockdown of PCSK9 results in increased recycling of LDL receptors on the surface of hepatocytes. Thus, the higher receptor density in the hepatocyte plasma membrane leads to enhanced LDL-C binding and reduced levels of circulating LDL-C [118]. Several clinical trials with larger numbers of patients have been performed. Most recently, the final results from three phase III studies, ORION-9 (heterozygous familial hypercholesterolemia), ORION-10 (artherosclerotic vascular disease), and ORION-11 (artherosclerotic vascular disease outside the USA) have been released. Collectively, 300 mg of inclisiran was administered at days 1, 90, and 270, and a > 60% reduction in PCSK9 levels as well as a durable reduction in LDL-C by 51% at days 510 or 540 were observed. Treatment was associated with lower levels of non-high-density lipoprotein cholesterol, total cholesterol, triglycerides, and apolipoprotein B [119, 120]. Participating patients are eligible for enrollment into an ongoing OKE (ORION-8) for monitoring the effects of inclisiran over 3 years. In Europe, Leqvio® received approval in late 2020 while the FDA rejected approval because of unresolved issues with inspection-related conditions at a third-party manufacturing facility. By mid-2021, Novartis had announced the transfer of the manufacturing of inclisiran to an Austria-based Novartis-owned facility and the complete response resubmission. Still, whether decreased PCSK9 and LDL-C levels will translate into improved cardiovascular outcomes remains to be seen, considering previous studies on the monoclonal anti-PCSK9 antibody evolocumab, which was able to reduce LDL-C without improving cardiovascular mortality. Indeed, in a pooled analysis of ORION-9, ORION-10, and ORION-11, only a 2.5 decrease in major cardiovascular events upon inclisiran treatment was found [121].
6.4 Lumasiran and Nedosiran
Two siRNA drugs/drug candidates, nedosiran and lumasiran, exist for the treatment of primary hyperoxaluria (PH), a very rare inherited disease characterized by an accumulation of hepatic glyoxylate. Primary hyperoxaluria can be sub-classified into three groups, based on different gene mutations. By lactate dehydrogenase (LDH), glyoxylate is metabolized to oxalate, which is filtered by the kidneys and leads to recurrent calcium oxalate kidney stones. One siRNA strategy aimed at the knockdown of LDH for inhibiting this final enzymatic step in oxalate production ([122]; see Sect. 7). Another target is glycolate oxidase, which catalyzes the conversion of glycolate to the main oxalate precursor glyoxylate [123].
Lumasiran (ALN-GO1, Oxlumo®; Alnylam) targets glycolate oxidase and has already gained FDA approval for PH type 1 (PH1) in November 2020. [124]. Again, it uses Alnylam’s ESC and GalNAc platform technologies. Its safety and efficacy was assessed in children aged > 6 years and adults with PH1 (ILLUMINATE-A; [125]; [126]). As a primary endpoint, a decrease of urinary oxalate levels over months 3–6 was defined. Indeed, 65.4% and 53.5% reductions relative to baseline and placebo, respectively, were seen. All secondary endpoints were met as well, including the proportion of patients reaching normalization or near normalization of urinary oxalate levels and the reduction in plasma oxalate. Most ILLUMINATE-A patients rolled over into the extension period of ILLUMINATE-B (infants and young children aged < 6 years) or ILLUMINATE-C (adults with advanced PH1), which are both ongoing.
6.5 Vutrisiran
Vutrisiran (Alnylam) is an siRNA drug that targets TTR for treating the same disease as patisiran, i.e., hereditary variant transthyretin amyloidosis. However, it is based on Alnylam’s enhanced stabilization chemistry and third-generation GalNAc-siRNA delivery platform [127,128,129]. It was granted orphan drug designation by the FDA and the EMA, as well as a Fast Track designation in the USA. Clinical phase III studies on the safety and efficacy are ongoing in both patients with hereditary transthyretin-mediated with polyneuropathy (HELIOS-A; vutrisiran vs patisiran) or cardiomyopathy (HELIOS-B; vutrisiran vs placebo). Based on positive 9-month results from the HELIOS-A study, vutrisiran received FDA approval in June 2022.
7 The Future: Other siRNAs in Other Diseases
Beyond the first siRNA therapeutics having reached the market, several others are in early or late-stage clinical trials (Table 3).
7.1 Metabolic Diseases
Nedosiran (DCR-PHXC; Dicerna Therapeutics) is another drug developed for the treatment of PH (see above). It specifically inhibits the expression of the major LDH isoform in the liver. As this is the final common step in the synthesis of oxalate, LDH knockdown is also efficient for treating the other hyperoxaluria subtypes, i.e., PH2 and PH3, and may be more potent than GO targeting by lumasiran. As a GalNAc-siRNA conjugate, it is subcutaneously injected once per month. Data from the multi-dose open-label PHYOX3 trial showed a sustained long-term urinary oxalate reduction in patients with PH1 and PH2, reaching normal or near-normal ranges [130]. In agreement with preclinical data in mice and non-human primates [122] as well as the absence of liver-specific adverse effects in natural LDHA-deficient patients [131], the drug was generally well tolerated. Clinical studies were also expanded towards patients with PH3 and early end-stage kidney disease. Recent data from a phase III study in infants and young children demonstrate a rapid sustained reduction in the spot urinary oxalate-to-creatinine ratio and plasma oxalate as well as acceptable safety in patients aged < 6 years with PH1 [130].
Cemdisiran is another liver-targeted GalNAc-siRNA drug that leads to the knockdown of the complement 5 (C5) protein. It is under development for the treatment of life-threatening rare complement-mediated diseases [132], including paroxysmal nocturnal hemoglobinuria, immunoglobulin A nephropathy, atypical hemolytic uremic syndrome, and generalized myasthenia gravis. The reduction in circulating C5 protein levels inhibits the activity of the terminal complement pathway. Consequently, the formation of the membrane attack complex and the release of the C5a anaphylatoxin are prevented [133, 134]. This knockdown also covers a point mutant of the C5 gene observed in some patients with paroxysmal nocturnal hemoglobinuria. Early clinical studies revealed a rapid, robust, and sustained C5 suppression maintained up to 13 months following single and multiple doses, supporting the further evaluation of cemdisiran as a single therapeutic or combined with complement inhibitor antibodies [135]. A phase II study evaluating its effects on proteinuria in adults with immunoglobulin A nephropathy is ongoing.
7.2 Hematology
Hemophilia A and B are the indications for fitusiran (ALN-AT3), a GalNAc-siRNA conjugate targeting the SERPINC1 mRNA. By thus reducing antithrombin production and increasing thrombin generation, fitusiran corrects the coagulation imbalance and prevents the bleeding phenotype [136,137,138]. In phase I and phase II trials in patients with or without inhibitors, fitusiran caused a very profound, dose-dependent antithrombin reduction [139, 140]. Phase III trials have been completed or are still ongoing. Given its profound and durable effects on hemostasis, independent of the presence of an inhibitor and reversible with antithrombin, the absence of anti-drug antibody formation (as opposed to immunotherapeutics), the favorable dosing regimen (once-monthly subcutaneous injection), and the safety profile with limited adverse effects, fitusiran emerges as a promising drug.
7.3 Infectious Diseases
Viral diseases are addressed as well in siRNA drug development programs. The siRNA drug RG6346 mediates selective knockdown of the viral hepatitis B surface antigen, which is required for the hepatitis V virus lifecycle, in liver hepatocytes. Results from a phase I trial presented in November 2020 showed four-monthly doses to lead to a substantial and durable reduction in hepatitis B surface antigen levels, lasting up to 1 year after the last dose. A phase II study of RG6346 alone or in combination with other investigational drugs/approved long-term hepatitis B virus treatments was initiated in March 2021 (NCT04225715). More recently, the first siRNA drug targeting coronavirus disease 2019 (COVID-19) was tested in clinical studies as well. In vitro, an siRNA targeting severe acute respiratory syndrome coronavirus 2 RNA-dependent RNA polymerase was identified as most efficient for inhibition of viral replication. This siR-7 was formulated with a peptide dendrimer (KK-46). In a Syrian hamster model for severe acute respiratory syndrome coronavirus 2 infection, a significant reduction in virus titers and lung inflammation was observed upon exposing animals to inhalation of siR-7-EM/KK-46 [17]. Based on these data, the efficacy and safety of MIR 19 were tested in a phase II, randomized, controlled, multi-center study in patients with symptomatic moderate COVID-19 (NCT05184127).
7.4 Oncology
An example from oncology is NU-0129, an siRNA targeting the cancer-promoting Bcl2L12 gene in glioblastoma. For delivery, brain-penetrant RNAi-based spherical nucleic acids are employed, consisting of gold nanoparticle cores covalently conjugated with radially oriented and densely packed siRNAs [141]. A single-arm, open-label, phase 0, first-in-human study in patients with recurrent glioblastoma demonstrated gold enrichment in the tumors (tumor cells, tumor-associated endothelium, macrophages) and a significant reduction in Bcl2L12 protein levels, with no significant treatment-related toxicity [141].
Mutated KRAS is the most prominent oncogenic driver in many cancers. An siRNA targeting the G12D mutant of KRAS has been formulated in a biodegradable polymeric matrix for sustained local release (LODER; Local Drug EluteR) [142]. Notably, a currently ongoing study in patients with locally advanced pancreatic cancer employs the combination of this siRNA with established chemotherapy based on gemcitabine or nab-paclitaxel [143].
7.5 Ocular Diseases
Several siRNA drug developments have looked into eye diseases. These include glaucoma, age-related macular degeneration (AMD), dry eye disease (dry eye syndrome), diabetic macular edema (DME), and various inherited retinal diseases. The first siRNA drug for treating ocular pathologies was bevasiranib in 2004. Bevasiranib was developed for targeting vascular endothelial growth factor (VEGF) in the treatment of DME and AMD. After promising phase I and phase II results on safety and efficacy [144], however, a phase III trial was terminated early because of a lack of efficacy in visual loss reduction (NCT00499590), and another phase III trial was withdrawn [145].
Going in the same direction, Sirna-027 was developed for targeting the VEGF receptor VEGFR1 in patients with AMD. Clinical studies demonstrated a significant improvement in best corrected visual acuity (BCVA); however, no correlation between the BCVA changes and the administered dose was observed. A phase II clinical trial was discontinued after recognizing off-target effects associated with TLR3 activation and the inability to achieve the desired visual acuity objective [146, 147].
The choroidal neovascularization resulting from AMD, diabetic retinopathy, and macular edema has been addressed by PF-04523655, an siRNA targeting the hypoxia-inducible gene RTP801 whose upregulation leads to neuronal cell death. Notably, its mechanism of action is thus independent of VEGF-targeted therapies. The phase II DEGAS trial in patients with visual loss due to DME, however, revealed only marginal efficacy when compared with focal/grid laser photocoagulation. The small improvement in BCVA and increasing patient discontinuation rates led again to the early termination of the trial. In contrast, the phase II MONET trial on PF-04523655 for the treatment of neovascular AMD revealed BCVA improvement. This was particularly so when combining PF-04523655 with the anti-VEGF drug ranibizumab. The results from a further phase II clinical trial (MATISSE; NCT01445899) for evaluating the safety, tolerability, and pharmacokinetic profile in patients with DME following dose escalation, as a monotherapy or in combination with ranibizumab versus ranibizumab alone have not been published yet.
Glaucoma as the leading cause of irreversible blindness worldwide. The main risk factor for glaucoma, increased intraocular pressure, is an imbalance between aqueous humour production and outflow. The β-adrenergic receptor 2 located in the eye has been shown to be involved in aqueous humour secretion [148]. Consequently, SYL040012 (Bamosiran), an siRNA targeting β-adrenergic receptor 2, has been developed. A phase II study in patients with glaucoma or elevated intraocular pressure demonstrated a statistically significant reduction in intraocular pressure at one dose level, with the drug being locally and systemically well tolerated [149].
Another indication of siRNAs in ocular diseases is dry eye syndrome, a multifactorial disease characterized by tear film disruption and damage of the ocular surface. Tivanisiran (SYL1001) targets the capsaicin receptor TRPV1 (transient receptor potential cation channel subfamily V member 1), which is involved in inflammatory response modulation and pain stimuli transmission. As a nociceptive transducer expressed in ocular tissues, it is critically involved in mediating ocular pain [150]. Clinical trials demonstrated tivanisiran to be efficient and safe upon topical application. Ocular pain and conjunctival hyperemia in dry eye syndrome were reduced [151].
7.6 Other Diseases
Acute kidney injury (AKI) affects up to 30% of patients undergoing cardiac surgery and thus contributes to morbidity and mortality. Reduced renal perfusion and ischemia reperfusion injury are the main mediators of the mechanisms contributing to postoperative AKI development. On the cellular level, ischemia reperfusion injury leads to cell injury and death. As the transcription factor p53 activates genes responsible for growth arrest or cell death after exposure to ischemia reperfusion injury, teprasiran has been developed for p53 knockdown. It has been evaluated for the prevention of AKI and its consequences in high-risk patients undergoing cardiac surgery [71]. In a phase II study, the incidence, severity, and duration of early AKI in high-risk patients undergoing cardiac surgery were found to be significantly reduced after teprasiran administration [152] and provided the basis for a phase III study. Beyond AKI, teprasiran has also been evaluated for reducing the incidence and severity of delayed graft function with kidney allografts (NCT02610296).
8 The Outlook I: siRNA Delivery Issues Revisited
The Human Genome Project as well as substantial progress in technologies such as next-generation sequencing and other major breakthroughs in the last decades have provided a wealth of information about the human genome and the pathophysiological role of genetic factors in various diseases [153, 154]. This has greatly extended the portfolio of possible targets for therapeutic intervention; however, many of those prove to be essentially un-druggable by small-molecule compounds or biologicals at the protein level. Not surprisingly, RNAs are thus considered as important targets for specific therapy. More recently, major advances also include a growing insight into the pivotal (patho-)physiological roles of different non-coding RNAs such as miRNAs, circRNAs, long non-coding RNAs, and others. Again, the straightforward, if not only possible approach for therapeutic intervention is RNA-based drugs. Thus, RNAs have proven to be much more versatile than initially thought, being targets as well as drugs. Twenty years after the discovery of RNAi, this has paved the way towards the first siRNA therapeutics.
Currently, five siRNA drugs have been approved (patisiran, givosiran, inclisiran, lumasiran, vutrisiran) and several others are in late stages of phase III clinical trials. Notably, there is a clear focus on hepatocyte delivery, with GalNAc being the most popular platform for siRNA bioconjugation and delivery. For good reasons, the vast majority of all siRNA drugs in clinical trials are based on GalNAc conjugates: GalNAc bioconjugates are comparatively straightforward to synthesize and administer (subcutaneously), they show a favorable biocompatibility/toxicity profile, and very high efficacy. Among the approved siRNA drugs, nanoparticles (LNPs) are only used in the case of patisiran, which certainly has to be considered as a landmark in the development of siRNA therapeutics. However, in the light of other hereditary transthyretin-mediated-targeted siRNA drugs, including vutrisiran, the future of patisiran may be limited.
Despite this obvious success in translating siRNA drugs into the clinics, the limitation to liver hepatocytes must be kept in mind. While some major advances have been made regarding central nervous system, ocular, or renal siRNA delivery, a ‘second siRNA drug breakthrough’ may be required with regard to other extra-hepatic siRNA delivery platforms and siRNA applications. Here, LNPs as well as other nanoparticles may play pivotal roles. The major requirements remain the same: efficacy in siRNA delivery, safety, biocompatibility/biodegradability as well as addressing issues regarding production, standardization, and approval as multi-component systems.
In parallel, siRNA conjugation partners other than GalNAc may offer potential for clinical translation: RGD for binding to integrins or folate for binding to the folate receptor on cancer cells; glucagon-like peptide 1 for binding its receptor on pancreatic beta cells; transferrin for binding to the transferrin receptor protein 1, to only name a few. In a more general way, antibodies binding to their cell-specific surface targets can be considered as an interesting conjugation partner. Notably, this approach has eventually reached the clinics in the form of immunotoxins such as gemtuzumab ozogamicin (Mylotarg®) or trastuzumab emtansin (Kadcyla®).
Beyond delivery to the desired organ, a major bottleneck is also the intracellular processing, as siRNAs are dependent on an exclusively intracellular mode of action. In this regard, it is of relevance that, upon cellular internalization, only a very small proportion of siRNAs escapes the endosomal/lysosomal system, with the vast majority of the siRNA molecules being either degraded or recycled back to the surface. In the case of LNPs, only a 1–2% fraction of the internalized siRNAs was found to be actually released into the cytoplasm [6, 155]. This highlights the requirement for nanoparticles capable of increasing the release of siRNAs from the endosomes/lysosomes. Alternative strategies are the co-delivery of endosome-disrupting small-molecule drugs [156], or the avoidance of the endosomal/lysosomal pathway in favor of the endosome-Golgi-ER pathway, which may lead to higher siRNA efficacy as shown in a recent study [157]. Approaches may also include looking into natural delivery systems such as extracellular vesicles (exosomes, others) or extracellular vesicle components. Indeed, in preclinical studies, exosomes have already been used for short-hairpin RNA delivery in pancreatic tumor-bearing mice [158] and the extracellular vesicle modification of nanoparticles such as polyethylenimine/siRNA complexes has been found to enhance nanoparticle efficacy and to efficiently deliver siRNAs to mouse tumor xenografts [159]. Mesenchymal stromal cell-derived exosomes are already used in a clinical study for KRAS G12D siRNA delivery, for the treatment of metastatic pancreatic ductal adenocarcinoma in patients with a KrasG12D mutation (NCT03608631).
Still, nanoparticles such as LNPs are comparatively large in size (often ~100 nm and above) and their extravasation from the bloodstream may therefore be limited to fenestrated endothelia. While this makes them optimal for the liver, but also solid tumors because of the enhanced permeability and retention effect [160], the preference for fenestrated tissues may limit their clinical use in other diseases. Other drawbacks of nanoparticles such as LNPs include that it is often necessary to administer them intravenously, making it a comparatively complex process that is unfavorable for patients. Additionally, the possible toxicity of their excipients may require pre-treatment of patients with anti-inflammatory drugs as seen in the case of patisiran. This is not so in the case of bioconjugates; however, they are limited by their dependency on suitable surface markers for binding and internalization. Clearly, there will not be a one-fits-all solution for all desired therapeutic siRNA applications.
8.1 The Outlook II: siRNA Drugs Emerging as One Standard Modality in Pharmacotherapy
Upon resolving the still existing problems regarding siRNA delivery, RNAi drugs will also reach the market for treating other pathologies. Beyond the obvious and already discussed aspect of being able to target otherwise un-druggable genes, they may well turn out to be easier to develop than small-molecule inhibitors, which, by acting on the protein level, require a high level of structural precision and improvement, and are therefore more challenging and complex to develop. At the same time, siRNA production is easier and cheaper as compared with antibodies, thus making them competitive also price wise, and siRNA therapeutics may come with advantages regarding the mode of administration and the dosing period. This is highlighted for example by the direct comparison of the anti-PCSK9 monoclonal antibody evolocumab (Repatha®) with the PCSK9-targeting siRNA drug inclisiran: the latter is substantially cheaper to synthesize and relies on less frequent administrations (only three subcutaneous injections at days 1, 90, and 270 in the above-mentioned clinical studies; [161]).
Beyond the exploration of oncogenes as obvious targets in oncology, as is already done for example by knockdown of mutant KRAS, the treatment of many other diseases is feasible as well. This may also include neurodegeneration, as recently demonstrated in preclinical studies on Alzheimer’s disease (siRN-30), or viral diseases. Needless to say that the COVID-19 pandemic and the development of RNA-based vaccines have further boosted the development and public visibility of RNA-based drugs. The treatment of viral infections such as COVID-19 based on siRNA-mediated knockdown of crucial target genes is feasible as well, as already shown on the preclinical level. Other diseases that are sometimes based on rather subtle mutations will become druggable as well. Beyond siRNAs, small RNA drugs may well also include miRNAs for miRNA replacement or antimiRNAs for miRNA inhibition.
9 The Conclusion: More siRNA Drugs Ahead
More than two decades after the discovery of RNAi, siRNA drugs have opened novel avenues towards innovative therapies in many diseases. Despite several major challenges and issues, including siRNA delivery and side effects, which have delayed the clinical translation of this entirely new class of pharmacological compounds, several siRNA drugs have received approval for clinical use and several more siRNAs are in late-stage clinical studies. An important step will be the further exploration of siRNA drugs beyond the liver, still emphasizing the need for appropriate delivery strategies. The almost unlimited availability of siRNAs against any target gene also highlights the need for identifying optimal targets. Furthermore, considering siRNA combinations or siRNAs against non-coding RNAs highlights that the potential of siRNA drugs has only partially been explored so far.
References
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11.
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8.
Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–9.
Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33.
Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–9.
Wittrup A, Ai A, Liu X, Hamar P, Trifonova R, Charisse K, et al. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat Biotechnol. 2015;33:870–6.
Setten RL, Rossi JJ, Han SP. The current state and future directions of RNAi-based therapeutics. Nature Rev Drug Disc. 2019;18:421–46.
Dixon SJ, Stockwell BR. Identifying druggable disease-modifying gene products. Curr Opin Chem Biol. 2009;13:549–55.
Sullenger BA, Nair S. From the RNA world to the clinic. Science. 2016;352:1417–20.
Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118.
Rafiee A, Riazi-Rad F, Havaskary M, Nuri F. Long noncoding RNAs: regulation, function and cancer. Biotechnol Genet Eng Rev. 2018;34:153–80.
Friedrich M, Wiedemann K, Reiche K, Puppel SH, Pfeifer G, Zipfel I, et al. The role of lncRNAs TAPIR-1 and -2 as diagnostic markers and potential therapeutic targets in prostate cancer. Cancers (Basel). 2020;12:1122.
Wilson JA, Richardson CD. Future promise of siRNA and other nucleic acid based therapeutics for the treatment of chronic HCV. Infect Disord Drug Targets. 2006;6:43–56.
DeVincenzo J, Lambkin-Williams R, Wilkinson T, Cehelsky J, Nochur S, Walsh E, et al. A randomized, double-blind, placebo-controlled study of an RNAi-based therapy directed against respiratory syncytial virus. Proc Natl Acad Sci U S A. 2010;107:8800–5.
Tompkins SM, Lo CY, Tumpey TM, Epstein SL. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci U S A. 2004;101:8682–6.
Li BJ, Tang Q, Cheng D, Qin C, Xie FY, Wei Q, et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat Med. 2005;11:944–51.
Khaitov M, Nikonova A, Shilovskiy I, Kozhikhova K, Kofiadi I, Vishnyakova L, et al. Silencing of SARS-CoV-2 with modified siRNA-peptide dendrimer formulation. Allergy. 2021;76:2840–54.
Idris A, Davis A, Supramaniam A, Acharya D, Kelly G, Tayyar Y, et al. A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. Mol Ther. 2021;29:2219–26.
Saify Nabiabad H, Amini M, Demirdas S. Specific delivering of RNAi using Spike’s aptamer-functionalized lipid nanoparticles for targeting SARS-CoV-2: a strong anti-Covid drug in a clinical case study. Chem Biol Drug Des. 2022;99:233–46.
Chang YC, Yang CF, Chen YF, Yang CC, Chou YL, Chou HW, et al. A siRNA targets and inhibits a broad range of SARS-CoV-2 infections including Delta variant. EMBO Mol Med. 2022;14(4):e15298.
Friedrich M, Pfeifer G, Binder S, Aigner A, Vollmer Barbosa P, Makert GR, et al. Selection and validation of siRNAs preventing uptake and replication of SARS-CoV-2. Front Bioeng Biotechnol. 2022;10: 801870.
Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–88.
Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci U S A. 2002;99:6047–52.
Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20:500–5.
Fakhr E, Zare F, Teimoori-Toolabi L. Precise and efficient siRNA design: a key point in competent gene silencing. Cancer Gene Ther. 2016;23:73–82.
Marques JT, Williams BR. Activation of the mammalian immune system by siRNAs. Nat Biotechnol. 2005;23:1399–405.
Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, Liang XJ. Therapeutic siRNA: state of the art. Signal Transduct Target Ther. 2020;5:101.
Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol. 2020;20:537–51.
Puthenveetil S, Whitby L, Ren J, Kelnar K, Krebs JF, Beal PA. Controlling activation of the RNA-dependent protein kinase by siRNAs using site-specific chemical modification. Nucleic Acids Res. 2006;34:4900–11.
Lester SN, Li K. Toll-like receptors in antiviral innate immunity. J Mol Biol. 2014;426:1246–64.
Amarzguioui M, Prydz H. An algorithm for selection of functional siRNA sequences. Biochem Biophys Res Commun. 2004;316:1050–8.
Ui-Tei K, Naito Y, Nishi K, Juni A, Saigo K. Thermodynamic stability and Watson-Crick base pairing in the seed duplex are major determinants of the efficiency of the siRNA-based off-target effect. Nucleic Acids Res. 2008;36:7100–9.
Safari F, Rahmani Barouji S, Tamaddon AM. Strategies for improving siRNA-induced gene silencing efficiency. Adv Pharm Bull. 2017;7:603–9.
Birmingham A, Anderson E, Sullivan K, Reynolds A, Boese Q, Leake D, et al. A protocol for designing siRNAs with high functionality and specificity. Nat Protoc. 2007;2:2068–78.
Jagla B, Aulner N, Kelly PD, Song D, Volchuk A, Zatorski A, et al. Sequence characteristics of functional siRNAs. RNA. 2005;11:864–72.
Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–30.
Tafer H. Bioinformatics of siRNA design. Methods Mol Biol. 2014;1097:477–90.
Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16.
Hardin CC, Watson T, Corregan M, Bailey C. Cation-dependent transition between the quadruplex and Watson-Crick hairpin forms of d(CGCG3GCG). Biochemistry. 1992;31:833–41.
Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015;4: e252.
Patel M, Bartom ET, Paudel B, Kocherginsky M, O’Shea KL, Murmann AE, et al. Identification of the toxic 6mer seed consensus for human cancer cells. Sci Rep. 2022;12:5130.
Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K, et al. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006;12:1188–96.
Shibahara S, Mukai S, Nishihara T, Inoue H, Ohtsuka E, Morisawa H. Site-directed cleavage of RNA. Nucleic Acids Res. 1987;15:4403–15.
Lu ZJ, Mathews DH. OligoWalk: an online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 2008;36:W104–8.
Lu ZJ, Mathews DH. Efficient siRNA selection using hybridization thermodynamics. Nucleic Acids Res. 2008;36:640–7.
Tafer H, Ameres SL, Obernosterer G, Gebeshuber CA, Schroeder R, Martinez J, et al. The impact of target site accessibility on the design of effective siRNAs. Nat Biotechnol. 2008;26:578–83.
Ding Y, Chan CY, Lawrence CE. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–41.
Shao Y, Chan CY, Maliyekkel A, Lawrence CE, Roninson IB, Ding Y. Effect of target secondary structure on RNAi efficiency. RNA. 2007;13:1631–40.
Sayers EW, Beck J, Bolton EE, Bourexis D, Brister JR, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021;49:D10–7.
Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, et al. Ensembl 2021. Nucleic Acids Res. 2021;49:D884–91.
Lee BT, Barber GP, Benet-Pagès A, Casper J, Clawson H, Diekhans M, et al. The UCSC Genome Browser database: 2022 update. Nucleic Acids Res. 2022;50:D1115–22.
Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug Discov Today. 2008;13:842–55.
Khvorova A, Watts JK. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol. 2017;35:238–48.
Shen X, Corey DR. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018;46:1584–600.
Inoue H, Hayase Y, Imura A, Iwai S, Miura K, Ohtsuka E. Synthesis and hybridization studies on two complementary nona(2′-O-methyl)ribonucleotides. Nucleic Acids Res. 1987;15:6131–48.
Dowler T, Bergeron D, Tedeschi AL, Paquet L, Ferrari N, Damha MJ. Improvements in siRNA properties mediated by 2′-deoxy-2′-fluoro-beta-D-arabinonucleic acid (FANA). Nucleic Acids Res. 2006;34:1669–75.
Kenski DM, Butora G, Willingham AT, Cooper AJ, Fu W, Qi N, et al. siRNA-optimized modifications for enhanced in vivo activity. Mol Ther Nucleic Acids. 2012;1: e5.
Kauppinen S, Vester B, Wengel J. Locked nucleic acid (LNA): high affinity targeting of RNA for diagnostics and therapeutics. Drug Discov Today Technol. 2005;2:287–90.
Bennett CF. Therapeutic antisense oligonucleotides are coming of age. Annu Rev Med. 2019;70:307–21.
Crooke ST, Vickers TA, Liang XH. Phosphorothioate modified oligonucleotide-protein interactions. Nucleic Acids Res. 2020;48:5235–53.
Huang Y, Hong J, Zheng S, Ding Y, Guo S, Zhang H, Zhang X, Du Q, Liang Z. Elimination pathways of systemically delivered siRNA. Mol Ther. 2011;19:381–5.
Geselowitz DA, Neckers LM. Bovine serum albumin is a major oligonucleotide-binding protein found on the surface of cultured cells. Antisense Res Dev. 1995;5:213–7.
Shen W, De Hoyos CL, Migawa MT, Vickers TA, Sun H, Low A, Bell TA 3rd, et al. Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat Biotechnol. 2019;37:640–50.
Migawa MT, Shen W, Wan WB, Vasquez G, Oestergaard ME, Low A, et al. Site-specific replacement of phosphorothioate with alkyl phosphonate linkages enhances the therapeutic profile of gapmer ASOs by modulating interactions with cellular proteins. Nucleic Acids Res. 2019;47:5465–79.
Haraszti RA, Roux L, Coles AH, Turanov AA, Alterman JF, Echeverria D, et al. 5′-Vinylphosphonate improves tissue accumulation and efficacy of conjugated siRNAs in vivo. Nucleic Acids Res. 2017;45:7581–92.
Parmar R, Willoughby JL, Liu J, Foster DJ, Brigham B, Theile CS, et al. 5′-(E)-Vinylphosphonate: a stable phosphate mimic can improve the RNAi activity of siRNA-GalNAc conjugates. ChemBioChem. 2016;17:985–9.
Elkayam E, Parmar R, Brown CR, Willoughby JL, Theile CS, Manoharan M, Joshua-Tor L. siRNA carrying an (E)-vinylphosphonate moiety at the 5΄ end of the guide strand augments gene silencing by enhanced binding to human Argonaute-2. Nucleic Acids Res. 2017;45:5008.
Valenzuela RA, Suter SR, Ball-Jones AA, Ibarra-Soza JM, Zheng Y, Beal PA. Base modification strategies to modulate immune stimulation by an siRNA. ChemBioChem. 2015;16:262–7.
Phelps KJ, Ibarra-Soza JM, Tran K, Fisher AJ, Beal PA. Click modification of RNA at adenosine: structure and reactivity of 7-ethynyl- and 7-triazolyl-8-aza-7-deazaadenosine in RNA. ACS Chem Biol. 2014;9:1780–7.
Wu SY, Chen TM, Gmeiner WH, Chu E, Schmitz JC. Development of modified siRNA molecules incorporating 5-fluoro-2′-deoxyuridine residues to enhance cytotoxicity. Nucleic Acids Res. 2013;41:4650–9.
Demirjian S, Ailawadi G, Polinsky M, Bitran D, Silberman S, Shernan SK, et al. Safety and tolerability study of an intravenously administered small interfering ribonucleic acid (siRNA) post on-pump cardiothoracic surgery in patients at risk of acute kidney injury. Kidney Int Rep. 2017;2:836–43.
Solano EC, Kornbrust DJ, Beaudry A, Foy JW, Schneider DJ, Thompson JD. Toxicological and pharmacokinetic properties of QPI-1007, a chemically modified synthetic siRNA targeting caspase 2 mRNA, following intravitreal injection. Nucleic Acid Ther. 2014;24:258–66.
Adams D, Gonzalez-Duarte A, O’Riordan WD, Yang CC, Ueda M, Kristen AV, et al. Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N Engl J Med. 2018;379:11–21.
Sardh E, Harper P, Balwani M, Stein P, Rees D, Bissell DM, et al. Phase 1 trial of an RNA interference therapy for acute intermittent porphyria. N Engl J Med. 2019;380:549–58.
Balwani M, Sardh E, Ventura P, Peiró PA, Rees DC, Stölzel U, et al. Phase 3 trial of RNAi therapeutic givosiran for acute intermittent porphyria. N Engl J Med. 2020;382:2289–301.
Fitzgerald K, White S, Borodovsky A, Bettencourt BR, Strahs A, Clausen V, et al. A highly durable RNAi therapeutic inhibitor of PCSK9. N Engl J Med. 2017;376:41–51.
Liebow A, Li X, Racie T, Hettinger J, Bettencourt BR, Najafian N, et al. An investigational RNAi therapeutic targeting glycolate oxidase reduces oxalate production in models of primary hyperoxaluria. J Am Soc Nephrol. 2017;28:494–503.
Wooddell CI, Blomenkamp K, Peterson RM, Subbotin VM, Schwabe C, Hamilton J, et al. Development of an RNAi therapeutic for alpha-1-antitrypsin liver disease. JCI Insight. 2020;5: e135348.
Butler AA, Price CA, Graham JL, Stanhope KL, King S, Hung YH, et al. Fructose-induced hypertriglyceridemia in rhesus macaques is attenuated with fish oil or ApoC3 RNA interference. J Lipid Res. 2019;60:805–18.
Javanbakht H, Mueller H, Walther J, Zhou X, Lopez A, Pattupara T, et al. Liver-targeted anti-HBV single-stranded oligonucleotides with locked nucleic acid potently reduce HBV gene expression in vivo. Mol Ther Nucleic Acids. 2018;11:441–54.
Kim YJ. Computational siRNA design considering alternative splicing. Methods Mol Biol. 2010;623:81–92.
Sun G, Rossi JJ. Problems associated with reporter assays in RNAi studies. RNA Biol. 2009;6:406–11.
Vervaeke P, Borgos SE, Sanders NN, Combes F. Regulatory guidelines and preclinical tools to study the biodistribution of RNA therapeutics. Adv Drug Deliv Rev. 2022;184: 114236.
Gao S, Dagnaes-Hansen F, Nielsen EJ, Wengel J, Besenbacher F, Howard KA, et al. The effect of chemical modification and nanoparticle formulation on stability and biodistribution of siRNA in mice. Mol Ther. 2009;17:1225–33.
Rajappa M, Saxena P, Kaur J. Ocular angiogenesis: mechanisms and recent advances in therapy. Adv Clin Chem. 2010;50:103–21.
Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12:967–77.
Wicki A, Witzigmann D, Balasubramanian V, Huwyler J. Nanomedicine in cancer therapy: challenges, opportunities, and clinical applications. J Control Release. 2015;200:138–57.
Kim B, Park JH, Sailor MJ. Rekindling RNAi therapy: materials design requirements for in vivo siRNA delivery. Adv Mater. 2019;31:e1903637.
Aigner A. Perspectives, issues and solutions in RNAi therapy: the expected and the less expected. Nanomedicine (Lond). 2019;14:2777–82.
Dong Y, Siegwart DJ, Anderson DG. Strategies, design, and chemistry in siRNA delivery systems. Adv Drug Deliv Rev. 2019;144:133–47.
Dowdy SF. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol. 2017;35:222–9.
Khorev O, Stokmaier D, Schwardt O, Cutting B, Ernst B. Trivalent, Gal/GalNAc-containing ligands designed for the asialoglycoprotein receptor. Bioorg Med Chem. 2008;16:5216–31.
Rajeev KG, Nair JK, Jayaraman M, Charisse K, Taneja N, O’Shea J, et al. Hepatocyte-specific delivery of siRNAs conjugated to novel non-nucleosidic trivalent N-acetylgalactosamine elicits robust gene silencing in vivo. ChemBioChem. 2015;16:903–8.
Prakash TP, Graham MJ, Yu J, Carty R, Low A, Chappell A, et al. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014;42:8796–807.
Springer AD, Dowdy SF. GalNAc-siRNA Conjugates: leading the way for delivery of RNAi therapeutics. Nucleic Acid Ther. 2018;28:109–18.
Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–6.
Sato Y, Hatakeyama H, Sakurai Y, Hyodo M, Akita H, Harashima H. A pH-sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. J Control Release. 2012;163:267–76.
Akinc A, Zumbuehl A, Goldberg M, Leshchiner ES, Busini V, Hossain N, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26:561–9.
Jayaraman M, Ansell SM, Mui BL, Tam YK, Chen J, Du X, et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem Int Ed Engl. 2012;51:8529–33.
Mui BL, Tam YK, Jayaraman M, Ansell SM, Du X, Tam YY, et al. Influence of polyethylene glycol lipid desorption rates on pharmacokinetics and pharmacodynamics of siRNA lipid nanoparticles. Mol Ther Nucleic Acids. 2013;2: e139.
Whitehead KA, Matthews J, Chang PH, Niroui F, Dorkin JR, Severgnini M, et al. In vitro-in vivo translation of lipid nanoparticles for hepatocellular siRNA delivery. ACS Nano. 2012;6:6922–9.
Cedillo I, Chreng D, Engle E, Chen L, McPherson AK, Rodriguez AA. Synthesis of 5′-GalNAc-conjugated oligonucleotides: a comparison of solid and solution-phase conjugation strategies. Molecules. 2017;22:1356.
Al Shaer D, Al Musaimi O, Albericio F, de la Torre BG. 2018 FDA tides harvest. Pharmaceuticals (Basel). 2019;12:52.
Wood H. FDA approves patisiran to treat hereditary transthyretin amyloidosis. Nat Rev Neurol. 2018;14:570.
Hoy SM. Patisiran: first global approval. Drugs. 2018;78:1625–31.
Zhang X, Goel V, Robbie GJ. Pharmacokinetics of patisiran, the first approved RNA interference therapy in patients with hereditary transthyretin-mediated amyloidosis. J Clin Pharmacol. 2019;60:573–85.
Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T, et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med. 2013;369:819–29.
Solomon SD, Adams D, Kristen A, Grogan M, González-Duarte A, Maurer MS, et al. Effects of patisiran, an RNA interference therapeutic, on cardiac parameters in patients with hereditary transthyretin-mediated amyloidosis. Circulation. 2019;139:431–43.
Schmidt HH, Wixner J, Planté-Bordeneuve V, Muñoz-Beamud F, Lladó L, Gillmore JD, et al. Patisiran treatment in patients with hereditary transthyretin-mediated amyloidosis with polyneuropathy after liver transplantation. Am J Transplant. 2022;22:1646–57.
Scott LJ. Givosiran: first approval. Drugs. 2020;80:335–9.
Al Shaer D, Al Musaimi O, Albericio F, de la Torre BG. 2019 FDA TIDES (peptides and oligonucleotides) harvest. Pharmaceuticals (Basel). 2020;13:40.
Wang B, Rudnick S, Cengia B, Bonkovsky HL. Acute hepatic porphyrias: review and recent progress. Hepatol Commun. 2019;3:193–206.
Yasuda M, Gan L, Chen B, Kadirvel S, Yu C, Phillips JD, et al. RNAi-mediated silencing of hepatic Alas1 effectively prevents and treats the induced acute attacks in acute intermittent porphyria mice. Proc Natl Acad Sci U S A. 2014;111:7777–82.
Chan A, Liebow A, Yasuda M, Gan L, Racie T, Maier M, et al. Preclinical development of a subcutaneous ALAS1 RNAi therapeutic for treatment of hepatic porphyrias using circulating RNA quantification. Mol Ther Nucleic Acids. 2015;4: e263.
Agarwal S, Simon AR, Goel V, Habtemariam BA, Clausen VA, Kim JB, et al. Pharmacokinetics and pharmacodynamics of the small interfering ribonucleic acid, givosiran, in patients with acute hepatic porphyria. Clin Pharmacol Ther. 2020;108:63–72.
Syed YY. Givosiran: a review in acute hepatic porphyria. Drugs. 2021;81:841–8.
Ventura P, Bonkovsky HL, Gouya L, Aguilera-Peiró P, Montgomery Bissell D, Stein PE, et al. Efficacy and safety of givosiran for acute hepatic porphyria: 24-month interim analysis of the randomized phase 3 ENVISION study. Liver Int. 2021;42:161–72.
Dyrbuś K, Gąsior M, Penson P, Ray KK, Banach M. Inclisiran: new hope in the management of lipid disorders? J Clin Lipidol. 2020;14:16–27.
Raal FJ, Kallend D, Ray KK, Turner T, Koenig W, Wright RS, et al. Inclisiran for the treatment of heterozygous familial hypercholesterolemia. N Engl J Med. 2020;382:1520–30.
Ray KK, Wright RS, Kallend D, Koenig W, Leiter LA, Raal FJ, et al. Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N Engl J Med. 2020;382:1507–19.
Bamji AN. Do PCSK9 inhibitors do anything more than reduce LDL cholesterol? BMJ Clin Res. 2020;368: m1159.
Lai C, Pursell N, Gierut J, Saxena U, Zhou W, Dills M, et al. Specific inhibition of hepatic lactate dehydrogenase reduces oxalate production in mouse models of primary hyperoxaluria. Mol Ther. 2018;26:1983–95.
Dutta C, Avitahl-Curtis N, Pursell N, Larsson Cohen M, Holmes B, Diwanji R, et al. Inhibition of glycolate oxidase with Dicer-substrate siRNA reduces calcium oxalate deposition in a mouse model of primary hyperoxaluria type 1. Mol Ther. 2016;24:770–8.
Scott LJ. Keam SJ. Lumasiran: first approval. Drugs. 2021;81:277–82.
Garrelfs SF, Frishberg Y, Hulton SA, Koren MJ, O’Riordan WD, Cochat P, et al. Lumasiran, an RNAi therapeutic for primary hyperoxaluria type 1. N Engl J Med. 2021;384:1216–26.
Frishberg Y, Deschênes G, Groothoff JW, Hulton SA, Magen D, Harambat J, et al. Phase 1/2 study of lumasiran for treatment of primary hyperoxaluria type 1: a placebo-controlled randomized clinical trial. Clin J Am Soc Nephrol. 2021;16:1025–36.
Foster DJ, Brown CR, Shaikh S, Trapp C, Schlegel MK, Qian K, et al. Advanced siRNA designs further improve in vivo performance of GalNAc-siRNA conjugates. Mol Ther. 2018;26:708–17.
Janas MM, Zlatev I, Liu J, Jiang Y, Barros SA, Sutherland JE, et al. Safety evaluation of 2′-deoxy-2′-fluoro nucleotides in GalNAc-siRNA conjugates. Nucleic Acids Res. 2019;47:3306–20.
Habtemariam BA, Karsten V, Attarwala H, Goel V, Melch M, Clausen VA, et al. Single-dose pharmacokinetics and pharmacodynamics of transthyretin targeting N-acetylgalactosamine-small interfering ribonucleic acid conjugate, vutrisiran, in healthy subjects. Clin Pharmacol Ther. 2021;109:372–82.
Hoppe B, Koch A, Cochat P, Garrelfs SF, Baum MA, Groothoff JW, et al. Safety, pharmacodynamics, and exposure-response modeling results from a first-in-human phase 1 study of nedosiran (PHYOX1) in primary hyperoxaluria. Kidney Int. 2021;101:626–34.
Kanno T, Sudo K, Maekawa M, Nishimura Y, Ukita M, Fukutake K. Lactate dehydrogenase M-subunit deficiency: a new type of hereditary exertional myopathy. Clin Chim Acta. 1988;173:89–98.
Noris M, Remuzzi G. Overview of complement activation and regulation. Semin Nephrol. 2013;33:479–92.
Rother RP, Rollins SA, Mojcik CF, Brodsky RA, Bell L. Discovery and development of the complement inhibitor eculizumab for the treatment of paroxysmal nocturnal hemoglobinuria. Nat Biotechnol. 2007;25:1256–64.
Woodruff TM, Nandakumar KS, Tedesco F. Inhibiting the C5–C5a receptor axis. Mol Immunol. 2011;48:1631–42.
Badri P, Jiang X, Borodovsky A, Najafian N, Kim J, Clausen VA, et al. Pharmacokinetic and pharmacodynamic properties of cemdisiran, an RNAi therapeutic targeting complement component 5, in healthy subjects and patients with paroxysmal nocturnal hemoglobinuria. Clin Pharmacokinet. 2021;60:365–78.
Sehgal A, Barros S, Ivanciu L, Cooley B, Qin J, Racie T, et al. An RNAi therapeutic targeting antithrombin to rebalance the coagulation system and promote hemostasis in hemophilia. Nat Med. 2015;21:492–7.
Pasi KJ, Rangarajan S, Georgiev P, Mant T, Creagh MD, Lissitchkov T, et al. Targeting of antithrombin in hemophilia A or B with RNAi therapy. N Engl J Med. 2017;377:819–28.
Machin N, Ragni MV. An investigational RNAi therapeutic targeting antithrombin for the treatment of hemophilia A and B. J Blood Med. 2018;9:135–40.
Pasi KJ, Lissitchkov T, Mamonov V, Mant T, Timofeeva M, Bagot C, et al. Targeting of antithrombin in hemophilia A or B with investigational siRNA therapeutic fitusiran: results of the phase 1 inhibitor cohort. J Thromb Haemost. 2021;19:1436–46.
Ragni MV, Georgiev P, Mant T, Creagh MD, Lissitchkov T, Bevan D, et al. Fitusiran, an investigational RNAi therapeutic targeting antithrombin for the treatment of hemophilia: updated results from a phase 1 and phase 1/2 extension study in patients without inhibitors. Blood. 2016;128:2572.
Kumthekar P, Ko CH, Paunesku T, Dixit K, Sonabend AM, Bloch O, et al. A first-in-human phase 0 clinical study of RNA interference-based spherical nucleic acids in patients with recurrent glioblastoma. Science Transl Med. 2021;13:eabb3945.
Golan T, Khvalevsky EZ, Hubert A, Gabai RM, Hen N, Segal A, et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget. 2015;6:24560–70.
Varghese AM, Ang C, Dimaio CJ, Javle MM, Gutierrez M, Yarom N, et al. A phase II study of siG12D-LODER in combination with chemotherapy in patients with locally advanced pancreatic cancer (PROTACT). J Clin Oncol. 2020;38:TPS4672-TPS.
Singerman L. Combination therapy using the small interfering RNA bevasiranib. Retina. 2009;29:S49-50.
Garba AO, Mousa SA. Bevasiranib for the treatment of wet, age-related macular degeneration. Ophthalmol Eye Dis. 2010;2:75–83.
Kleinman ME, Yamada K, Takeda A, Chandrasekaran V, Nozaki M, Baffi JZ, et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–7.
Cho WG, Albuquerque RJ, Kleinman ME, Tarallo V, Greco A, Nozaki M, et al. Small interfering RNA-induced TLR3 activation inhibits blood and lymphatic vessel growth. Proc Natl Acad Sci USA. 2009;106:7137–42.
Lu LJ, Tsai JC, Liu J. Novel pharmacologic candidates for treatment of primary open-angle glaucoma. Yale J Biol Med. 2017;90:111–8.
Gonzalez V, Palumaa K, Turman K, Muñoz FJ, Jordan J, García J, et al. Phase 2 of bamosiran (SYL040012), a novel RNAi based compound for the treatment of increased intraocular pressure associated to glaucoma. Invest Ophthalmol Vis Sci. 2014;55:564.
Moreno-Montañés J, Bleau AM, Jimenez AI. Tivanisiran, a novel siRNA for the treatment of dry eye disease. Exp Opin Invest Drugs. 2018;27:421–6.
Benitez-Del-Castillo JM, Moreno-Montañés J, Jiménez-Alfaro I, Muñoz-Negrete FJ, Turman K, Palumaa K, et al. Safety and efficacy clinical trials for SYL1001, a novel short interfering RNA for the treatment of dry eye disease. Invest Ophthalmol Vis Sci. 2016;57:6447–54.
Thielmann M, Corteville D, Szabo G, Swaminathan M, Lamy A, Lehner LJ, et al. Teprasiran, a small interfering RNA, for the prevention of acute kidney injury in high-risk patients undergoing cardiac surgery: a randomized clinical study. Circulation. 2021;144:1133–44.
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921.
Lekka E, Hall J. Noncoding RNAs in disease. FEBS Lett. 2018;592:2884–900.
Sahay G, Querbes W, Alabi C, Eltoukhy A, Sarkar S, Zurenko C, et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat Biotechnol. 2013;31:653–8.
Du Rietz H, Hedlund H, Wilhelmson S, Nordenfelt P, Wittrup A. Imaging small molecule-induced endosomal escape of siRNA. Nat Commun. 2020;11:1809.
Qiu C, Han HH, Sun J, Zhang HT, Wei W, Cui SH, et al. Regulating intracellular fate of siRNA by endoplasmic reticulum membrane-decorated hybrid nanoplexes. Nat Commun. 2019;10:2702.
Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503.
Zhupanyn P, Ewe A, Buch T, Malek A, Rademacher P, Muller C, et al. Extracellular vesicle (ECV)-modified polyethylenimine (PEI) complexes for enhanced siRNA delivery in vitro and in vivo. J Control Release. 2020;319:63–76.
Fang J, Nakamura H, Maeda H. The EPR effect: unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Adv Drug Deliv Rev. 2011;63:136–51.
Sheridan C. PCSK9-gene-silencing, cholesterol-lowering drug impresses. Nat Biotechnol. 2019;37:1385–7.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
Open Access funding enabled and organized by Projekt DEAL.
Conflicts of interest/Competing interests
The authors have no conflicts of interest to declare.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Authors’ contributions
Both authors contributed to the outlining, writing, and proofreading of this article.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Friedrich, M., Aigner, A. Therapeutic siRNA: State-of-the-Art and Future Perspectives. BioDrugs 36, 549–571 (2022). https://doi.org/10.1007/s40259-022-00549-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40259-022-00549-3