Abstract
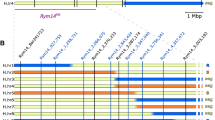
Soil-borne barley yellow mosaic virus disease, caused by different strains of Barley yellow mosaic virus (BaYMV) and Barley mild mosaic virus (BaMMV), is one of the most important diseases of winter barley (Hordeum vulgare L.) in Europe and East Asia. The recessive resistance gene rym11 located in the centromeric region of chromosome 4HL is effective against all so far known strains of BaMMV and BaYMV in Germany. In order to isolate this gene, a high-resolution mapping population (10,204 meiotic events) has been constructed. F2 plants were screened with co-dominant flanking markers and segmental recombinant inbred lines (RILs) were tested for resistance to BaMMV under growth chamber and field conditions. Tightly linked markers were developed by exploiting (1) publicly available barley EST sequences, (2) employing barley synteny to rice, Brachypodium distachyon and sorghum and (3) using next-generation sequencing data of barley. Using this approach, the genetic interval was efficiently narrowed down from the initial 10.72 % recombination to 0.074 % recombination. A marker co-segregating with rym11 was developed providing the basis for gene isolation and efficient marker-assisted selection.


Similar content being viewed by others
References
Andersen JR, Lübberstedt T (2003) Functional markers in plants. Trends Plant Sci 8:554–560
Bauer E, Weyen J, Schiemann A, Graner A, Ordon F (1997) Molecular mapping of novel resistance genes against barley mild mosaic virus (BMMV). Theor Appl Genet 95:1263–1269
Bevan M, Waugh R (2007) Meeting report: applying plant genomics to crop improvement. Genome Biol 8:302
Clark MF, Adams AN (1977) Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J Gen Virol 34:475–483
Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N et al (2009) Development and implementation of high-throughput SNP genoyping in barley. BMC Genomics 10:582
Comadran J, Ramsay L, MacKenzie K, Hayes P, Close TJ, Muehlbauer G, Stein N, Waugh R (2010) Patterns of polymorphism and linkage disequilibrium in cultivated barley. Theor Appl Genet 122:523–531
Dorokhov DB, Klocke E (1997) A rapid and economic technique for RAPD analysis of plant genomes. Russ J Gen 33:358–365
Duran C, Appleby N, Vardy M, Imelfort M, Edwardsand D, Batley J (2009) Single nucleotide polymorphism discovery in barley using autoSNPdb. Plant Biotechnol J 7:326–333
Dvorák J, Luo MC, Yang ZL (1998) Restriction fragment length polymorphism and divergence in the genomic regions of high and low recombination in self-fertilizing and cross-fertilizing aegilops species. Genetics 148:423–434
Fraser RSS (1990) The genetics of resistance to plant viruses. Annu Rev Phytopathol 28:179–200
Friedt W, Ordon F (2007) Molecular markers for gene pyramiding and disease resistance breeding in barley. In: Varshney R, Tuberosa R (eds) Genomics assisted crop improvement. Springer, Berlin, pp 81–101
Friedt W, Humbroich K, Ordon F (2008) Genetic diversity of resistance against barley yellow mosaic virus complex in barley. In: Proceedings of the 10th International barley genetics symposium, Alexandria, Egypt, vol 6, pp 7–12
Gottwald S, Stein N, Börner A, Sasaki T, Graner A (2004) The gibberellic-acid insensitive dwarfing gene sdw3 of barley is located on chromosome 2HS in a region that shows high colinearity with rice chromosome 7L. Mol Genet Genomics 271:426–436
Götz R, Friedt W (1993) Resistance to the barley yellow mosaic virus complex—differential genotypic reactions and genetics of BaMMV-resistance of barley (Hordeum vulgare L.). Plant Breeding 111:125–131
Graner A, Streng S, Kellermann A, Schiemann A, Bauer E, Waugh R, Pellio B, Ordon F (1999) Molecular mapping and fine structure of the rym5 locus encoding resistance to different strains of the Barley Yellow Mosaic Virus Complex. Theor Appl Genet 98:285–290
Habekuss A, Kühne T, Krämer I, Rabenstein F, Ehrig F, Ruge-Wehling B, Huth W, Ordon F (2008) Identification of barley mild mosaic virus isolates in Germany breaking rym5 resistance. J Phytopathol 156:36–41
Hanemann A, Schweizer GF, Cossu R, Wicker T, Röder MS (2009) Fine mapping, physical mapping and development of diagnostic markers for the Rrs2 scald resistance gene in barley. Theor Appl Genet 119:1507–1522
Hariri D, Meyer M, Le Gouis J, Bahrman N, Fouchard M, Forget C, Andre A (2000) Characterisation of BaYMV and BaMMV pathotypes in France. Eur J Plant Pathol 106:365–372
Hariri D, Meyer M, Prud’homme H (2003) Characterization of a new barley mild mosaic virus pathotype in France. Eur J Plant Pathol 109:921–928
He P, Li JZ, Zheng XW, Shen LS, Lu CF, Chen Y, Zhu LH (2001) Comparison of molecular linkage maps and agronomic trait loci between DH and RIL populations derived from the same rice cross. Crop Sci 41:1240–1246
Hearnden PR, Eckermann PJ, McMichael GL, Hayden MJ, Eglinton JK, Chalmers KJ (2007) A genetic map of 1,000 SSR and DArT markers in a wide barley cross. Theor Appl Genet 115:383–391
Hofinger BJ, Russell JR, Bass CG, Baldwin T, Dos Reis M, Hedley PE, Li Y, Macaulay M, Waugh R, Hammond-Kosack KE, Kanyuka K (2011) An exceptionally high nucleotide and haplotype diversity and a signature of positive selection for the eIF4E resistance gene in barley are revealed by allele mining and phylogenetic analyses of natural populations. Mol Ecol. doi:10.1111/j.1365-294X.2011.05201.x
Huth W (1991) Verbreitung der Gelbmosaikvirosen BaYMV, BaMMV, und BaYMV-2 und Screening von Gerstensorten auf Resistenz gegenüber BaYMV-2. Nachrichtenbl Deut Pflanzenschutzd 43:233–237
Kai H, Takata K, Tsukazaki M, Furusho M, Baba T (2012) Molecular mapping of Rym17, a dominant and rym18 a recessive barley yellow mosaic virus (BaYMV) resistance genes derived from Hordeum vulgare L. Theor Appl Genet 124:577–583
Kanyuka K, Ward E, Adams MJ (2003) Polymyxa graminis and the cereal viruses it transmits: a research challenge. Mol Plant Pathol 4:393–406
Kanyuka K, McGrann G, Alhudaib K, Hariri D, Adams MJ (2004) Biological and sequence analysis of a novel European isolate of Barley mild mosaic virus that overcomes the barley rym5 resistance gene. Arch Virol 149:1469–1480
Kanyuka K, Druka A, Caldwell DG, Tymon A, McCallum N, Waugh R, Adams MJ (2005) Evidence that the recessive bymovirus resistance locus rym4 in barley corresponds to the eukaryotic translation initiation factor 4E gene. Mol Plant Pathol 6:449–458
Kota R, Varshney RK, Prasad M, Zhang H, Stein N, Graner A (2008) EST-derived single nucleotide polymorphism markers for assembling genetic and physical maps of the barley genome. Funct Integr Genomics 8:223–233
Kühne T (2009) Soil-borne viruses affecting cereals: known for long but still a threat. Virus Res 141:174–183
Künzel G, Korzun L, Meister A (2000) Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics 154:397–412
Langridge P (2005) Molecular breeding of wheat and barley. In: Tuberosa R, Phillips RL, Gale M (eds) In the wake of the double helix: from the green revolution to the gene revolution. Avenue media, Bologna, pp 279–286
Lebel EG, Masson J, Bogucki A, Paszkowski J (1993) Stress-induced intrachromosomal recombination in plant somatic cells. Proc Natl Acad Sci USA 90:422–426
López-Moya JJ, Garcia JA (2008) Potyviruses. In: Mahy BWJ, van Regenmortel M (eds) Encyclopedia of virology, 3rd edn. Elsevier, Oxford, pp 313–322
Maule AJ, Caranta C, Boulton MI (2007) Sources of natural resistance to plant viruses: status and prospects. Mol Plant Pathol 8:223–231
Mayer KFX, Martis M, Hedley PE, Kimková H, Liu H, Morris JA, Steuernagel B, Taudien S, Roessner S, Gundlach H, Kubaláková M, Suchánková P, Murat F, Felder M, Nussbaumer T, Graner A, Salse J, Endo T, Sakai H, Tanaka T, Itoh T, Sato K, Platzer M, Matsumoto T, Scholz U, Doleqel J, Waugh R, Stein N (2011) Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23:1249–1263
Nissan-Azzouz F, Graner A, Friedt W, Ordon F (2005) Fine-mapping of the BaMMV, BaYMV-1 and BaYMV-2 resistance of barley (Hordeum vulgare) accession PI1963. Theor Appl Genet 110:212–218
Ordon F, Götz R, Friedt W (1993) Genetic stocks resistant to barley yellow mosaic viruses (BaMMV, BaYMV, BaYMV-2) in Germany. Barley Genet Newsl 22:46–49
Ordon F, Friedt W, Scheurer K, Pellio B, Werner K, Neuhaus G, Huth W, Habekuss A, Graner A (2004) Molecular markers in breeding for virus resistance in barley. J Appl Genet 45:145–159
Palloix A, Ordon F (2011) Advanced breeding for virus resistance in plants. In: Caranta C, Aranda MA, Tepfer M, Lopez-Moya JJ (eds) Recent advances in plant virology. Caister Academic Press, UK, pp 195–218
Pecinka A, Rosa M, Schikora A, Berlinger M, Hirt H, Luschnig C, Mittelsten Scheid O (2009) Transgenerational stress memory is not a general response in Arabidopsis. PLoS One 4:5202
Pellio B, Streng S, Bauer E, Stein N, Perovic D, Schiemann A, Friedt W, Ordon F, Graner A (2005) High-resolution mapping of the Rym4/Rym5 locus conferring resistance to the barley yellow mosaic virus complex (BaMMV, BaYMV, BaYMV-2) in barley (Hordeum vulgare ssp. vulgare L.). Theor Appl Genet 110:283–293
Perovic D, Stein N, Zhang H, Drescher A, Prasad M, Kota R, Kopahnke D, Graner A (2004) An integrated approach for comparative mapping in rice and barley with special reference to the Rph16 resistance locus. Funct Integr Genomics 4:74–83
Perovic D, Tiffin P, Douchkov D, Bäumlein H, Graner A (2007) An integrated approach for the comparative analysis of a multigene family: the nicotianamine synthase genes of barley. Funct Integr Genomics 7:169–179
Perovic D, Zorić D, Milovanović M, Prodanović S, Yan Y, Janković S, Šurlan-Momirović G (2009) Hordein gene dose effects in triploid endosperm of barley (Hordeum vulgare L.). Genetika 41:271–287
Petes TD (2001) Meiotic recombination hot spots and cold spots. Nat Rev Genet 2:360–369
Provvidenti R, Hampton RO (1992) Sources of resistance to viruses in the Potyviridae. Arch Virol Suppl 5:189–211
Ramsay L, Macaulay M, degli Ivanissevich S, MacLean K, Cardle L, Fuller J, Edwards KJ, Tuvesson S, Morgante M, Massari A, Maestri E, Marmiroli N, Sjakste T, Ganal M, Powell W, Waugh R (2000) A simple sequence repeat-based linkage map of barley. Genetics 156:1997–2005
Revers F, Le Gall OT, Candresse T, Maule AJ (1999) New advances in understanding the molecular biology of plant/potyvirus interactions. Mol Plant Microbe Interact 12:367–376
Robaglia C, Caranta C (2006) Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci 11:40–45
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Ruge B, Linz A, Pickering R, Proeseler G, Greif P, Wehling P (2003) Mapping of Rym14Hb, a gene introgressed from Hordeum bulbosum and conferring resistance to BaMMV and BaYMV in barley. Theor Appl Genet 107:965–971
Ruge-Wehling B, Linz A, Habekuß A, Wehling P (2006) Mapping of Rym16 (Hb), the second soil-borne virus-resistance gene introgressed from Hordeum bulbosum. Theor Appl Genet 113:867–873
Sato K, Nankaku N, Takeda K (2009) A high-density transcript linkage map of barley derived from a single population. Heredity 103:110–117
Shahinnia F, Sayed-Tabatabaei BE, Sato K, Pourkheirandish M, Komatsuda T (2009) Mapping of QTL for intermedium spike on barley chromosome 4H using EST-based markers. Breeding Sci 59:383–390
Shahinnia F, Druka A, Franckowiak J, Morgante M, Robbie Waugh R, Stein N (2012) High resolution mapping of Dense spike-ar (dsp.ar) to the genetic centromere of barley chromosome 7H. Theor Appl Genet 124:373–384
Silvar C, Perovic D, Casas AM, Igartua E, Ordon F (2011) Development of a cost-effective pyrosequencing approach for SNP genotyping in barley. Plant Breeding 130:394–397
Silvar C, Perovic D, Scholz U, Casas AM, Igartua E, Ordon F (2012) Fine mapping and comparative genomics integration of two quantitative trait loci controlling resistance to powdery mildew in a Spanish barley landrace. Theor Appl Genet 124:49–62
Smilde WD, Haluskova J, Sasaki T, Graner A (2001) New evidence for the synteny of rice chromosome 1 and barley chromosome 3H from rice expressed sequence tags. Genome 44:361–367
Stein N, Graner A (2004) Map based gene isolation in cereal genomes. In: Gupta PK, Varshney RK (eds) Cereal genomics. Kluwer, Dordrecht, pp 331–360
Stein N, Herren G, Keller B (2001) A new DNA extraction method for high-throughput marker analysis in a large genome species such as Triticum aestivum. Plant Breed 120:354–356
Stein N, Perovic D, Kumlehn J, Pellio B, Stracke S, Streng S, Ordon F, Graner A (2005) The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J 42:912–922
Stein N, Prasad M, Scholz U, Thiel T, Zhang H, Wolf M, Kota R, Varshney RK, Perovic D, Graner A (2007) A 1000 loci transcript map of the barley Genome—new anchoring points for integrative grass genomics. Theor Appl Genet 114:823–839
Thiel T, Graner A, Waugh R, Grosse I, Close TJ, Stein N (2009) Evidence and evolutionary analysis of ancient whole-genome duplication in barley predating the divergence from rice. BMC Evol Biol 9:209
Thieme R, Rakosy-Tican E, Gavrilenko T, Antonova O, Schubert J, Nachtigall M, Heimbach U, Thieme T (2008) Novel somatic hybrids (Solanum tuberosum L. + Solanum tarnii) and their fertile BC1 progenies express extreme resistance to potato virus Y and late blight. Theor Appl Genet 116:691–700
Timpe U, Kühne T (1994) The complete nucleotide sequence of RNA2 of barley mild mosaic virus (BaMMV). Eur J Plant Pathol 100:233–241
Varshney RK, Marcel TC, Ramsay L, Russell J, Röder MS, Stein N, Waugh R, Langridge P, Niks RE, Graner A (2007) A high density barley microsatellite consensus map with 775 SSR loci. Theor Appl Genet 114:1091–1103
Vu GTH, Wicker T, Buchmann JP, Chandler PM, Matsumoto T, Graner A, Stein N (2010) Fine mapping and syntenic integration of the semi-dwarfing gene sdw3 of barley. Funct Integr Genomics 10:509–521
Wang ML, Barkley NA, Jenkins TM (2009) Microsatellite markers in plants and insects. Part I: applications of biotechnology. Genes Genomes Genomics 3:54–67
Werner K, Friedt W, Laubach E, Waugh R, Ordon F (2003) Dissection of resistance to soil-borne yellow-mosaic-inducing viruses of barley (BaMMV, BaYMV, BaYMV-2) in a complex breeders’ cross by means of SSRs and simultaneous mapping of BaYMV/BaYMV-2 resistance of var. ‘Chikurin Ibaraki 1’. Theor Appl Genet 106:1425–1432
Wu Y, Close TJ, Lonardi S (2008) On the accurate construction of consensus genetic maps. Comput Syst Bioinformatics Conf 7:285–296
Yeam I, Cavatorta JR, Ripoll DR, Kang BC, Jahn MM (2007) Functional dissection of naturally occurring amino acid substitutions in eIF4E that confers recessive potyvirus resistance in plants. Plant Cell 19:2913–2928
Acknowledgments
The authors thank Karin Ernst and Dörte Grau (JKI) for excellent technical assistance and Susanne König (IPK) for sequencing service. Furthermore, we thank the Federal Ministry of Education and Research (BMBF) for funding this project (FKZ 0314000D) within the BARLEX project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Hayes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lüpken, T., Stein, N., Perovic, D. et al. Genomics-based high-resolution mapping of the BaMMV/BaYMV resistance gene rym11 in barley (Hordeum vulgare L.). Theor Appl Genet 126, 1201–1212 (2013). https://doi.org/10.1007/s00122-013-2047-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-013-2047-3