Abstract
Key message
Marker-assisted backcrossing was used to generate pea NILs carrying individual or combined resistance alleles at main Aphanomyces resistance QTL. The effects of several QTL were successfully validated depending on genetic backgrounds.
Abstract
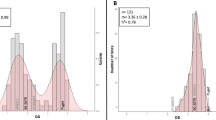
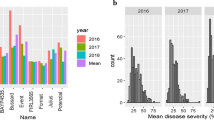
Quantitative trait loci (QTL) validation is an important and often overlooked step before subsequent research in QTL cloning or marker-assisted breeding for disease resistance in plants. Validation of QTL controlling partial resistance to Aphanomyces root rot, one of the most damaging diseases of pea worldwide, is of major interest for the future development of resistant varieties. The aim of this study was to validate, in different genetic backgrounds, the effects of various resistance alleles at seven main resistance QTL recently identified. Five backcross-assisted selection programs were developed. In each, resistance alleles at one to three of the seven main Aphanomyces resistance QTL were transferred into three genetic backgrounds, including two agronomically important spring (Eden) and winter (Isard) pea cultivars. The subsequent near-isogenic lines (NILs) were evaluated for resistance to two reference strains of the main A. euteiches pathotypes under controlled conditions. The NILs carrying resistance alleles at the major-effect QTL Ae-Ps4.5 and Ae-Ps7.6, either individually or in combination with resistance alleles at other QTL, showed significantly reduced disease severity compared to NILs without resistance alleles. Resistance alleles at some minor-effect QTL, especially Ae-Ps2.2 and Ae-Ps5.1, were also validated for their individual or combined effects on resistance. QTL × genetic background interactions were observed, mainly for QTL Ae-Ps7.6, the effect of which increased in the winter cultivar Isard. The pea NILs are a novel and valuable resource for further understanding the mechanisms underlying QTL and their integration in breeding programs.






Similar content being viewed by others
References
Ahmadi N, Albar L, Pressoir G, Pinel A, Fargette D, Ghesquiere A (2001) Genetic basis and mapping of the resistance to Rice yellow mottle virus. III. Analysis of QTL efficiency in introgressed progenies confirmed the hypothesis of complementary epistasis between two resistance QTLs. Theor Appl Genet 103:1084–1092
Balkunde S, Le HL, Lee HS, Kim DM, Kang JW, Ahn SN (2013) Fine mapping of a QTL for the number of spikelets per panicle by using near-isogenic lines derived from an interspecific cross between Oryza sativa and Oryza minuta. Plant Breed 132:70–76
Bates D, Maechler M, Bolker B, Walker S (2014) lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1-7. http://CRAN.R-project.org/package=lme4
Boutet G, Alves-Carvalho S, Falque M, Peterlongo P, Lhuillier E, Bouchez O, Lavaud C, Pilet-Nayel ML, Riviere N, Baranger A (2015) SNP discovery and genetic mapping using Genotyping by Sequencing of whole genome genomic DNA from a pea RIL population. BMC Genomics
Brouwer DJ, St Clair DA (2004) Fine mapping of three quantitative trait loci for late blight resistance in tomato using near isogenic lines (NILs) and sub-NILs. Theor Appl Genet 108:628–638
Castro P, Piston F, Madrid E, Millan T, Gil J, Rubio J (2010) Development of chickpea near-isogenic lines for Fusarium wilt. Theor Appl Genet 121:1519–1526
Chung CL, Longfellow JM, Walsh EK, Kerdieh Z, Van Esbroeck G, Balint-Kurti P, Nelson RJ (2010) Resistance loci affecting distinct stages of fungal pathogenesis: use of introgression lines for QTL mapping and characterization in the maize–Setosphaeria turcica pathosystem. BMC Plant Biol 10:103
Collard BC, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc B-Biol Sci 363:557–572
Delourme R, Piel N, Horvais R, Pouilly N, Domin C, Vallee P, Falentin C, Manzanares-Dauleux MJ, Renard M (2008) Molecular and phenotypic characterization of near isogenic lines at QTL for quantitative resistance to Leptosphaeria maculans in oilseed rape (Brassica napus L.). Theor Appl Genet 117:1055–1067
Didelot D, Chaillet I (1995) Relevance and interest of root disease prediction tests for pea crop in France. In: AEP (ed) 2nd Eur Conf Grain Legumes—improving production and utilisation of grain legumes. July 9–13th, Copenhagen- Denmark, p 150
Ding X, Li X, Xiong L (2011) Evaluation of near-isogenic lines for drought resistance QTL and fine mapping of a locus affecting flag leaf width, spikelet number, and root volume in rice. Theor Appl Genet 123:815–826
Duarte J, Riviere N, Baranger A, Aubert G, Burstin J, Cornet L, Lavaud C, Lejeune-Henaut I, Martinant JP, Pichon JP, Pilet-Nayel ML, Boutet G (2014) Transcriptome sequencing for high throughput SNP development and genetic mapping in Pea. BMC Genom 15:126
Falke KC, Frisch M (2011) Power and false-positive rate in QTL detection with near-isogenic line libraries. Heredity 106:576–584
Fujita D, Yoshimura A, Yasui H (2010) Development of near-isogenic lines and pyramided lines carrying resistance genes to green rice leafhopper (Nephotettix cincticeps Uhler) with the Taichung 65 genetic background in rice (Oryza sativa L.). Breed Sci 60:18–27
Graves S, Piepho HP, Selzer L, Dorai-Raj S (2012) multcompView: visualizations of paired comparisons. R package version 0.1-5. http://CRAN.R-project.org/package=multcompView
Gritton ET (1990) Registration of 5 root-rot resistant germplasm lines of processing pea. Crop Sci 30:1166–1167
Halekoh U, Højsgaard S (2013) pbkrtest: parametric bootstrap and Kenward Roger based methods for mixed model comparison. R package version 0.3-8. http://CRAN.R-project.org/package=pbkrtest
Hamon C, Baranger A, Coyne CJ, McGee RJ, Le Goff I, L’Anthoene V, Esnault R, Riviere JP, Klein A, Mangin P, McPhee KE, Roux-Duparque M, Porter L, Miteul H, Lesne A, Morin G, Onfroy C, Moussart A, Tivoli B, Delourme R, Pilet-Nayel ML (2011) New consistent QTL in pea associated with partial resistance to Aphanomyces euteiches in multiple French and American environments. Theor Appl Genet 123:261–281
Hamon C, Coyne CJ, McGee RJ, Lesne A, Esnault R, Mangin P, Herve M, Le Goff I, Deniot G, Roux-Duparque M, Morin G, McPhee KE, Delourme R, Baranger A, Pilet-Nayel ML (2013) QTL meta-analysis provides a comprehensive view of loci controlling partial resistance to Aphanomyces euteiches in four sources of resistance in pea. BMC Plant Biol 13:45
Hospital F (2003) Marker-assisted breeding. Blackwell Scientific Publishers, London
Hospital F (2005) Selection in backcross programmes. Philos Trans R Soc B-Biol Sci 360:1503–1511
Hospital F (2009) Challenges for effective marker-assisted selection in plants. Genetica 136:303–310
Hospital F, Charcosset A (1997) Marker-assisted introgression of quantitative trait loci. Genetics 147:1469–1485
John F, Sanford W (2011) An {R} Companion to Applied Regression, second edn. Sage. http://CRAN.R-project.org/package=car
Jones FR, Drechsler C (1925) Root rot of peas in the United States caused by Aphanomyces euteiches. J Agric Res 30:293–325
Kaeppler SM, Phillips RL, Kim TS (1993) Use of near-isogenic lines derived by backcrossing or selfing to map qualitative traits. Theor Appl Genet 87:233–237
Keller B, Manzanares C, Jara C, Lobaton JD, Studer B, Raatz B (2015) Fine-mapping of a major QTL controlling angular leaf spot resistance in common bean (Phaseolus vulgaris L.). Theor Appl Genet 128:813–826
Koester RP, Sisco PH, Stuber CW (1993) Identification of quantitative trait loci controlling days to flowering and plant height in 2 near-isogenic lines of maize. Crop Sci 33:1209–1216
Kraft JM (1992) Registration of 90-2079, 90-2131, and 90-2322 pea germplasms. Crop Sci 32:1076–1076
Lenth RV, Herve M (2015) lsmeans: least-squares means. R package version 2.18. http://CRAN.R-project.org/package=lsmeans
Li ZK, Yu SB, Lafitte HR, Huang N, Courtois B, Hittalmani S, Vijayakumar CHM, Liu GF, Wang GC, Shashidhar HE, Zhuang JY, Zheng KL, Singh VP, Sidhu JS, Srivantaneeyakul S, Khush GS (2003) QTL x environment interactions in rice. I. Heading date and plant height. Theor Appl Genet 108:141–153
Lockwood JL, Ballard JC (1960) Evaluation of pea introductions for resistance to aphanomyces and fusarium root rots. Michigan Quarterly Bulletin 42(4):704–713
Loridon K, McPhee K, Morin J, Dubreuil P, Pilet-Nayel ML, Aubert G, Rameau C, Baranger A, Coyne C, Lejeune-Henaut I, Burstin J (2005) Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor Appl Genet 111:1022–1031
Ma J, Yan GJ, Liu CJ (2011) Development of near-isogenic lines for a major QTL on 3BL conferring Fusarium crown rot resistance in hexaploid wheat. Euphytica 183:147–152
Malvick DK, Grau CR, Percich JA (1998) Characterization of Aphanomyces euteiches strains based on pathogenicity tests and random amplified polymorphic DNA analyses. Mycol Res 102:465–475
McGee RJ, Coyne CJ, Pilet-Nayel M-L, Moussart A, Tivoli B, Baranger A, Hamon C, Vandemark G, McPhee K (2012) Registration of pea germplasm lines partially resistant to aphanomyces root rot for breeding fresh or freezer pea and dry pea types. J Plant Regist 6:203
Moussart A, Wicker E, Duparque M, Rouxel F (2001) Development of an efficient screening test for pea resistance to Aphanomyces euteiches. In: AEP (ed) 4th Eur Conf Grain Legumes. July 8–12th, Cracow, Poland, pp 272–273
Moussart A, Onfroy C, Lesne A, Esquibet M, Grenier E, Tivoli B (2007) Host status and reaction of Medicago truncatula accessions to infection by three major pathogens of pea (Pisum sativum) and alfalfa (Medicago sativa). Eur J Plant Pathol 117:57–69
Moussart A, Even MN, Tivoli B (2008) Reaction of genotypes from several species of grain and forage legumes to infection with a French pea isolate of the oomycete Aphanomyces euteiches. Eur J Plant Pathol 122:321–333
Onfroy C, Tivoli B, Grünwald NJ, Pilet-Nayel ML, Baranger A, Andrivon D, Moussart A (2015) Aggressiveness and virulence of Aphanomyces euteiches isolates recovered from pea nurseries in the United States and France. Plant Pathol
Papavizas GC, Ayers WA (1974) Aphanomyces species and their root diseases in pea and sugarbeet. A review. US Dep Agric Tech Bull 1485:158
Pea G, Aung HH, Frascaroli E, Landi P, Pe ME (2013) Extensive genomic characterization of a set of near-isogenic lines for heterotic QTL in maize (Zea mays L.). BMC Genom 14:61
Pilet-Nayel ML, Muehlbauer FJ, McGee RJ, Kraft JM, Baranger A, Coyne CJ (2002) Quantitative trait loci for partial resistance to Aphanomyces root rot in pea. Theor Appl Genet 106:28–39
Pilet-Nayel ML, Muehlbauer FJ, McGee RJ, Kraft JM, Baranger A, Coyne CJ (2005) Consistent quantitative trait loci in pea for partial resistance to Aphanomyces euteiches isolates from the United States and France. Phytopathology 95:1287–1293
Pociecha E, Plazek A, Rapacz M, Niemczyk E, Zwierzykowski Z (2010) Photosynthetic activity and soluble carbohydrate content induced by the cold acclimation affect frost tolerance and resistance to microdochium nivale of androgenic festulolium genotypes. J Agron Crop Sci 196:48–54
R Development Core Team (2014) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Retrieved from http://www.R-project.org
Reyna N, Sneller CH (2001) Evaluation of marker-assisted introgression of yield QTL alleles into adapted soybean. Crop Sci 41:1317–1321
Richardson KL, Vales MI, Kling JG, Mundt CC, Hayes PM (2006) Pyramiding and dissecting disease resistance QTL to barley stripe rust. Theor Appl Genet 113:485–495
Salameh A, Buerstmayr M, Steiner B, Neumayer A, Lemmens M, Buerstmayr H (2011) Effects of introgression of two QTL for fusarium head blight resistance from Asian spring wheat by marker-assisted backcrossing into European winter wheat on fusarium head blight resistance, yield and quality traits. Mol Breed 28:485–494
Schmalenbach I, Korber N, Pillen K (2008) Selecting a set of wild barley introgression lines and verification of QTL effects for resistance to powdery mildew and leaf rust. Theor Appl Genet 117:1093–1106
Sindhu A, Ramsay L, Sanderson LA, Stonehouse R, Li R, Condie J, Shunmugam ASK, Liu Y, Jha AB, Diapari M, Burstin J, Aubert G, Tar’an B, Bett KE, Warkentin TD, Sharpe AG (2014) Gene-based SNP discovery and genetic mapping in pea. Theor Appl Genet 127:2225–2241
St Clair DA (2010) Quantitative disease resistance and quantitative resistance Loci in breeding. Annu Rev Phytopathol 48:247–268
Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203
Tayeh N, Aluome C, Falque M, Jacquin F, Klein A, Chauveau A, Bérard A, Houtin H, Rond C, Kreplak J, Boucherot K, Martin C, Baranger A, Pilet-Nayel M-L, Warkentin T, Brunel D, Marget P, Le Paslier M-C, Aubert G, Burstin J (2015) The GenoPea Infinium® BeadChip allowed the construction of a high-density high-resolution gene-based consensus genetic map in pea and provided an overview on the genome organization. Plant J (submitted)
Thabuis A, Palloix A, Servin B, Daubeze AM, Signoret P, Hospital F, Lefebvre V (2004) Marker-assisted introgression of 4 Phytophthora capsici resistance QTL alleles into a bell pepper line: validation of additive and epistatic effects. Mol Breed 14:9–20
Wicker E, Rouxel F (2001) Specific behaviour of French Aphanomyces euteiches Drechs. Populations for virulence and aggressiveness on pea, related to isolates from Europe, America and New Zealand. Eur J Plant Pathol 107:919–929
Wicker E, Moussart A, Duparque M, Rouxel F (2003) Further contributions to the development of a differential set of pea cultivars (Pisum sativum) to investigate the virulence of isolates of Aphanomyces euteiches. Eur J Plant Pathol 109:47–60
Xue S, Li G, Jia H, Lin F, Cao Y, Xu F, Tang M, Wang Y, Wu X, Zhang Z, Zhang L, Kong Z, Ma Z (2009) Marker-assisted development and evaluation of near-isogenic lines for scab resistance QTLs of wheat. Mol Breed 25:397–405
Yadav SK (2010) Cold stress tolerance mechanisms in plants. A review. Agron Sustain Dev 30:515–527
Yi M, Nwe KT, Vanavichit A, Chai-arree W, Toojinda T (2009) Marker assisted backcross breeding to improve cooking quality traits in Myanmar rice cultivar Manawthukha. Field Crop Res 113:178–186
Yu ZH, Mackill DJ, Bonman JM, Tanksley SD (1991) Tagging genes for blast resistance in rice via linkage to RFLP markers. Theor Appl Genet 81:471–476
Yu XM, Griffith M, Wiseman SB (2001) Ethylene induces antifreeze activity in winter rye leaves. Plant Physiol 126:1232–1240
Zhong ZZ, Wu WX, Wang HJ, Chen LP, Liu LL, Wang CM, Zhao ZG, Lu GW, Gao H, Wei XJ, Yu CY, Chen MJ, Shen YY, Zhang X, Cheng ZJ, Wang JL, Jiang L, Wan JM (2014) Fine mapping of a minor-effect QTL, DTH12, controlling heading date in rice by up-regulation of florigen genes under long-day conditions. Mol Breed 34:311–322
Zhou R, Zhu Z, Kong X, Huo N, Tian Q, Li P, Jin C, Dong Y, Jia J (2005) Development of wheat near-isogenic lines for powdery mildew resistance. Theor Appl Genet 110:640–648
Acknowledgments
This work was supported by a pre-doctoral fellowship (Clément Lavaud) from INRA, Département de Biologie et Amélioration des Plantes (France) and Brittany region (France). The SAMPOIS project from MAP (Ministère de l’Agriculture et de la Pêche, Paris, France) and Terres Univia (Paris, France) funded the NIL construction. The PeaMUST project, which received funding from the French Government managed by the Research National Agency (ANR) under the Investments for the Future call 2011 (ANR-11-BTBR-0002), funded the NIL evaluation studies. We acknowledge Solène Coëdel and Berline Fopa-Fomeju for having contributed to the technical work during NIL construction. We thank the genotyping GENTYANE and BIOGENOUEST platforms of Clermont-Ferrand and Rennes, France, for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
Authors declare that the described experiments comply with the French laws.
Additional information
Communicated by D. E. Mather.
C. Lavaud and A. Lesné contributed equally to the work described in the manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lavaud, C., Lesné, A., Piriou, C. et al. Validation of QTL for resistance to Aphanomyces euteiches in different pea genetic backgrounds using near-isogenic lines. Theor Appl Genet 128, 2273–2288 (2015). https://doi.org/10.1007/s00122-015-2583-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-015-2583-0