Abstract
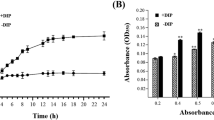
Vibrio splendidus is a ubiquitous pathogen that causes various diseases in aquaculture with a wide range of hosts. In our previous studies, we showed that l-glutamic acid was the optimal carbon source that could revive V. splendidus persister cells. In our present study, single cell observation under microscopy showed that V. splendidus could revive using l-glutamic acid as carbon source. A proteomic analysis was carried out to further illustrate the initial wake up of persister cells with l-glutamic acid. To collect the initially revived cells, SDS-PAGE was used to determine the revived time. The total proteins from the persister cells and the revived cells were analyzed using LC‒MS/MS. A total of 106 proteins, including 42 downregulated proteins and 64 upregulated proteins, were identified. GO analysis of the differentially expressed proteins (DEPs) showed that biological processes, including protein complex assembly, protein oligomerization, and arginine metabolism; cellular components, including extracellular membrane, plasma membrane and ribosome; and molecular functions, including the activities of arginine binding and structural constituent of ribosome, were enriched. KEGG analysis showed that lipopolysaccharide biosynthesis, porphyrin and chlorophyll metabolism, and peptidoglycan biosynthesis were upregulated, while the ribosome was downregulated. This is the first time to study the initial wake up of persister cells based on proteomic analysis, and the results revealed the main pathways involved in the early resuscitation of V. splendidus persister cells.






Similar content being viewed by others
References
Akiyama T, Williamson KS, Schaefer R, Pratt S, Chang CB, Franklin MJ (2017) Resuscitation of Pseudomonas aeruginosa from dormancy requires hibernation promoting factor (PA4463) for ribosome preservation. Proc Natl Acad Sci U S A 114(12):3204–3209
Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S (2004) Bacterial persistence as a phenotypic switch. Science 305(5690):1622–1625
Balchin D, Hayer-Hartl M, Hartl FU (2016) In vivo aspects of protein folding and quality control. Science 353(6294):aac4354
Chowdhury N, Wood TL, Martínez-Vázquez M, García-Contreras R, Wood TK (2016) DNA-crosslinker cisplatin eradicates bacterial persister cells. Biotechnol Bioeng 113(9):1984–1992
DiBiasio EC, Dickinson RA, Trebino CE, Ferreira CN, Morrison JJ, Camberg JL (2021) The stress-active cell division protein ZapE alters FtsZ filament architecture to facilitate division in Escherichia coli. Front Microbiol 12:733085
Epstein SS (2009) Microbial awakenings. Nature 457(7233):1083
Fisher RA, Gollan B, Helaine S (2017) Persistent bacterial infections and persister cells. Nat Rev Microbiol 15(8):453–464
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ (2012) STRING V9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 4:D808–D815
Goeminne LJE, Gevaert K, Clement L (2017) Experimental design and data-analysis in label-free quantitative LC/MS proteomics: a tutorial with MSqRob. J Proteom S1874–3919(17):30123–30129
Gollan B, Grabe G, Michaux C, Helaine S (2019) Bacterial persisters and infection: past, present, and progressing. Annu Rev Microbiol 73:359–385
Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13
Jiang G, Li Y, Li Y, Zhang W, Li C (2022) Selection of the amino acid and saccharide that increase the tetracycline susceptibility of Vibrio splendidus. Front Vet Sci 8:823332
Jolliffe IT, Cadima J (2016) Principal component analysis: a review andrecent developments. Philos Trans A A374:20150202
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240
Jubair M, Morris JG Jr (2012) Survival of Vibrio cholerae in nutrient-poor environments is associated with a novel “persister” phenotype. PLoS ONE 7(9):e45187
Keren I, Kaldalu N, Spoering A, Wang Y, Lewis K (2004) Persister cells and tolerance to antimicrobials. FEMS Microbiol Lett 230(1):13–18
Kim JS, Chowdhury N, Yamasaki R, Wood TK (2018a) Viable but non-culturable and persistence describe the same bacterial stress state. Environ Microbiol 20(6):2038–2048
Kim JS, Yamasaki R, Song S, Zhang W, Wood TK (2018b) Single cell observations show persister cells wake based on ribosome content. Environ Microbiol 20(6):2085–2098
Kwan BW, Chowdhury N, Wood TK (2015) Combating bacterial infections by killing persister cells with mitomycin C. Environ Microbiol 17(11):4406–4414
Li Y, Wood TK, Zhang W, Li C (2021) Vibrio splendidus persister cells induced by host coelomic fluids show a similar phenotype to antibiotic-induced counterparts. Environ Microbiol 23(9):5605–5620
Li Y, Dai F, Li YN, Liang WK, Li CH, Zhang WW (2022) Hfq, a global regulator contributes to the virulence of Vibrio splendidus AJ01. Aquaculture 546:737416
Liang W, Zhang W, Li C (2022) Vibrio splendidus virulence to Apostichopus japonicus is mediated by hppD through glutamate metabolism and flagellum assembly. Virulence 13(1):458–470
Marteyn BS, Karimova G, Fenton AK, Gazi AD, West N, Touqui L, Prevost MC, Betton JM, Poyraz O, Ladant D, Gerdes K, Sansonetti PJ, Tang CM (2014) ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. mBio 5(2):e00022–e00014
Megaw J, Gilmore BF (2017) Archaeal persisters: persister cell formation as a stress response in Haloferax volcanii. Front Microbiol 8:1589
Rahman M, Ismat F, Jiao L, Baldwin JM, Sharples DJ, Baldwin SA, Patching SG (2017) Characterisation of the DAACS family Escherichia coli glutamate/aspartate-proton symporter GltP using computational, chemical, biochemical and biophysical methods. J Membr Biol 250(2):145–162
Requena JM (2012) Stress response in microbiology. Caister Academic Press, Madrid
Schnier J, Nishi K (1988) Temperature-sensitive mutants with alterations in ribosomal protein L24 and isolation of intra- and extragenic suppressor mutants. Methods Enzymol 164:706–709
Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K (2006) Persisters: a distinct physiological state of E. coli. BMC Microbiol 6:53–61
Silva-Valenzuela CA, Lazinski DW, Kahne SC, Nguyen Y, Molina-Quiroz RC, Camilli A (2017) Growth arrest and a persister state enable resistance to osmotic shock and facilitate dissemination of Vibrio cholerae. ISME J 11(12):2718–2728
Sinha-Ray S, Ali A (2017) Mutation in flrA and mshA genes of Vibrio cholerae inversely involved in vps-independent biofilm driving bacterium toward nutrients in lake water. Front Microbiol 8:1770
Song S, Wood TK (2020) Persister cells resuscitate via ribosome modification by 23S rRNA pseudouridine synthase RluD. Environ Microbiol 22(3):850–857
Stapels DAC, Hill PWS, Westermann AJ, Fisher RA, Thurston TL, Saliba AE, Blommestein I, Vogel J, Helaine S (2018) Salmonella persisters undermine host immune defenses during antibiotic treatment. Science 362(6419):1156–1160
Van den Bergh B, Michiels JE, Wenseleers T, Windels EM, Boer PV, Kestemont D, De Meester L, Verstrepen KJ, Verstraeten N, Fauvart M, Michiels J (2016) Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat Microbiol 1:16020
van Ooijen MP, Jong VL, Eijkemans MJC, Heck AJR, Andeweg AC, Binai NA, van den Ham HJ (2018) Identification of differentially expressed peptides in high-throughput proteomics data. Brief Bioinform 19(5):971–981
Wiuff C, Zappala RM, Regoes RR, Garner KN, Baquero F, Levin BR (2005) Phenotypic tolerance: antibiotic enrichment of noninherited resistance in bacterial populations. Antimicrob Agents Chemother 49(4):1483–1494
Wood TK, Song S, Yamasaki R (2019) Ribosome dependence of persister cell formation and resuscitation. J Microbiol 57(3):213–219
Yamasaki R, Song S, Benedik MJ, Wood TK (2020) Persister cells resuscitate using membrane sensors that activate chemotaxis, lower cAMP levels, and revive ribosomes. iScience 23(1):100792
Zhang WW, Li CH (2021) Virulence mechanisms of vibrios belonging to the Splendidus clade as aquaculture pathogens, from case studies and genome data. Rev Aquacult 13:2004–2026
Zhang W, Yamasaki R, Song S, Wood TK (2019) Interkingdom signal indole inhibits Pseudomonas aeruginosa persister cell waking. J Appl Microbiol 127(6):1768–1775
Funding
This work was finally supported by the Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholar (LR20C190001), the National Natural Science Foundation of China (31972833), the Fundamental Research Funds for the Provincial Universities of Zhejiang (SJLZ2020001), and the K.C. Wong Magna Fund at Ningbo University.
Author information
Authors and Affiliations
Contributions
GJ: conducted the experiments, analyzed the data, and wrote the original manuscript. YL: conducted parts of the experiments and analyzed the data. JZ: conducted parts of the experiments. WL analyzed parts of the data. WD conceived and planned the research, and revised the manuscript. WZ: conceived and planned the research, supervised the research, revised the manuscript, and acquired funding.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest in this work.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, G., Li, Y., Zhang, J. et al. Proteomic analysis of the initial wake up of vibrio splendidus persister cells. World J Microbiol Biotechnol 39, 116 (2023). https://doi.org/10.1007/s11274-023-03559-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-023-03559-7