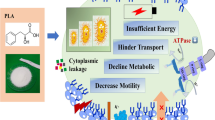
Abstract
Vibrio parahaemolyticus is a common pathogen in aquatic products, such as shellfishes. Laboratory-based simulated studies demonstrated that V. parahaemolyticus can tolerate high hydrostatic pressure (HHP) up to 20 MPa. However, the molecular mechanisms of high-pressure adaptation remain unclear. Herein, we analyzed the physiological changes and transcriptomic responses of V. parahaemolyticus ATCC 17,802 under HHP conditions to determine the possible survival mechanisms. Under HHP conditions, the morphology of V. parahaemolyticus was notably changed exhibiting the coccoid microbial cells. The transcriptome analysis revealed that there were 795 differentially expressed genes (DEGs) under the 20 MPa condition, including 406 upregulated DEGs and 389 downregulated DEGs. Most of the downregulated DEGs encoded proteins related to energy metabolism, such as citrate synthase (gltA), pyruvate kinase (pyk), and glyceraldehyde-3-phosphate dehydrogenase (gapA). Many of the upregulated DEGs encoded proteins related to adhesion and virulence factors, such as RNA polymerase σ factor (rpoE), L-threonine 3-dehydrogenase, and bacterial nucleotide signal c-di-GMP (WU75_RS02745 and WU75_RS07185). In our proposed mechanism model, V. parahaemolyticus responds to HHP stress through RNA polymerase σ factor RpoE. These findings indicate that V. parahaemolyticus cells may adopt a complex adaptation strategy to cope with HHP stress.
Key points
•The transcriptomic response of Vibrio parahaemolyticus under HHP conditions was studied for the first time.
•V. parahaemolyticus may adopt a complex adaptation strategy to cope with HHP stress.
•ToxRS and RpoE played an important role in sensing and responding the HHP signal.






Similar content being viewed by others
Data availability
Unfiltered, untrimmed, and paired-end RNA sequence data have been uploaded and made available in NCBI BioProject under the accession number PRJNA813199.
References
Aagesen AM, Phuvasate S, Su YC, Häse CC (2013) Persistence of Vibrio parahaemolyticus in the Pacific oyster, Crassostrea gigas, is a multifactorial process involving pili and flagella but not type III secretion systems or phase variation. Appl Environ Microbiol 79:3303–3305. https://doi.org/10.1128/AEM.00314-13
Abe F, Horikoshi K (2000) Tryptophan permease gene TAT2 confers high-pressure growth in Saccharomyces cerevisiae. Mol Cell Biol 20:8093–8102. https://doi.org/10.1128/MCB.20.21.8093-8102.2000
Aertsen A, Vanoirbeek K, De Spiegeleer P, Sermon J, Hauben K, Farewell A, Nyström T, Michiels CW (2004) Heat shock protein-mediated resistance to high hydrostatic pressure in Escherichia coli. Appl Environ Microbiol 70:2660–2666. https://doi.org/10.1128/AEM.70.5.2660-2666.2004
Aertsen A, Van Houdt R, Vanoirbeek K, Michiels CW (2004) An SOS response induced by high pressure in Escherichia coli. J Bacteriol 186:6133–6141. https://doi.org/10.1128/JB.186.18.6133-6141.2004
Alpas H, Lee J, Bozoglu F, Kaletunc G (2003) Evaluation of high hydrostatic pressure sensitivity of Staphylococcus aureus and Escherichia coli O157:H7 by differential scanning calorimetry. Int J Food Microbiol 87:229–237. https://doi.org/10.1016/S0168-1605(03)00066-7
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data
Asakura H, Kawamoto K, Murakami S, Tachibana M, Kurazono H, Makino S, Yamamoto S, Igimi S (2016) Ex vivo proteomics of Campylobacter jejuni 81-176 reveal that FabG affects fatty acid composition to alter bacterial growth fitness in the chicken gut. Res Microbiol 167:63–71. https://doi.org/10.1016/j.resmic.2015.10.001
Bai X, Nakatsu CH, Bhunia AK (2021) Bacterial biofilms and their implications in pathogenesis and food safety. Foods 10:2117. https://doi.org/10.3390/foods10092117
Bartlett D, Wright M, Yayanos AA, Silverman M (1989) Isolation of a gene regulated by hydrostatic pressure in a deep-sea bacterium. Nature 342:572–574. https://doi.org/10.1038/342572a0
Bekker M, De Vries S, Ter Beek A, Hellingwerf KJ, Teixeira de Mattos MJ (2009) Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd-II oxidase. J Bacteriol 191:5510–5517. https://doi.org/10.1128/JB.00562-09
Ben-Zvi AP, Goloubinoff P (2001) Mechanisms of disaggregation and refolding of stable protein aggregates by molecular chaperones. J Struct Biol 135:84–93. https://doi.org/10.1006/jsbi.2001.4352
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Champe PC, Harvey RA, Ferrier DR (2005) Biochemistry, 3rd edn. Lippincott Williams & Wilkins, Philadelphia, Pennsylvania
Chen SY, Jane WN, Chen YS, Wong HC (2009) Morphological changes of Vibrio parahaemolyticus under cold and starvation stresses. Int J Food Microbiol 129:157–165. https://doi.org/10.1016/j.ijfoodmicro.2008.11.009
Chen J, Liu H, Cai S, Zhang H (2019) Comparative transcriptome analysis of Eogammarus possjeticus at different hydrostatic pressure and temperature exposures. Sci Rep 9:1–10. https://doi.org/10.1038/s41598-019-39716-y
Chi E, Bartlett DH (1995) An rpoE-like locus controls outer membrane protein synthesis and growth at cold temperatures and high pressures in the deep-sea bacterium Photobacterium sp. strain SS9. Mol Microbiol 17:713–726. https://doi.org/10.1111/j.1365-2958.1995.mmi_17040713.x
Claret L, Calder SR, Higgins M, Hughes C (2003) Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Mol Microbiol 48:1349–1355. https://doi.org/10.1046/j.1365-2958.2003.03506.x
Coutard F, Lozach S, Pommepuy M, Hervio-Heath D (2007) Real-time reverse transcription-pcr for transcriptional expression analysis of virulence and housekeeping genes in viable but nonculturable Vibrio parahaemolyticus after recovery of culturability. Appl Environ Microbiol 73:5183–5189. https://doi.org/10.1128/AEM.02776-06
Coutard F, Crassous P, Droguet M, Gobin E, Colwell RR, Pommepuy M, Hervio-Heath D (2007) Recovery in culture of viable but nonculturable Vibrio parahaemolyticus: regrowth or resuscitation? ISME J 1:111–120. https://doi.org/10.1038/ismej.2007.1
DeLong EF, Yayanos AA (1985) Adaptation of the membrane lipids of a deep-sea bacterium to changes in hydrostatic pressure. Science 228:1101–1103. https://doi.org/10.1126/science.3992247
Enos-Berlage JL, Guvener ZT, Keenan CE, McCarter LL (2005) Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol Microbiol 55:1160–1182. https://doi.org/10.1111/j.1365-2958.2004.04453.x
Fu LL, Wang R, Wang Y, Lin J (2014) Proteomic identification of responsive proteins of Vibrio parahaemolyticus under high hydrostatic pressure. J Sci Food Agr 94:2630–2638. https://doi.org/10.1002/jsfa.6595
Gotoh K, Kodama T, Hiyoshi H, Izutsu K, Park KS, Dryselius R, Akeda Y, Honda T, Iida T (2010) Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants. PLoS ONE 5:e13365. https://doi.org/10.1371/journal.pone.0013365
Gu D, Guo M, Yang M, Zhang Y, Zhou X, Wang Q (2016) A σE-mediated temperature gauge controls a switch from LuxR-mediated virulence gene expression to thermal stress adaptation in Vibrio alginolyticus. PLoS Pathog 12:e1005645. https://doi.org/10.1371/journal.ppat.1005645
Haines-Menges B, Whitaker WB, Boyd EF (2014) Alternative sigma factor RpoE is important for Vibrio parahaemolyticus cell envelope stress response and intestinal colonization. Infect Immun 82:3667–3677. https://doi.org/10.1128/IAI.01854-14
Hammer BK, Bassler BL (2003) Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol 50:101–104. https://doi.org/10.1046/j.1365-2958.2003.03688.x
Ishii A, Oshima T, Sato T, Nakasone K, Mori H, Kato C (2005) Analysis of hydrostatic pressure effects on transcription in Escherichia coli by DNA microarray procedure. Extremophiles 9:65–73. https://doi.org/10.1007/s00792-004-0414-3
Jiang X, Chai TJ (1996) Survival of Vibrio parahaemolyticus at low temperatures under starvation conditions and subsequent resuscitation of viable, nonculturable cells. Appl Environ Microbiol 62:1300–1305. https://doi.org/10.1128/aem.62.4.1300-1305.1996
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Lee LH, Raghunath P (2018) Vibrionaceae diversity, multidrug resistance and management. Front Microbiol 9:563. https://doi.org/10.3389/fmicb.2018.00563
Letchumanan V, Chan KG, Pusparajah P, Saokaew S, Duangjai A, Goh BH, Mutalib NSA, Lee LH (2016) Insights into bacteriophage application in controlling Vibrio species. Front Microbiol 7:1114. https://doi.org/10.3389/fmicb.2016.01114
Li S, Xiao X, Li J, Luo J, Wang F (2006) Identification of genes regulated by changing salinity in the deep-sea bacterium Shewanella sp. WP3 using RNA arbitrarily primed PCR. Extremophiles 10:97–104. https://doi.org/10.1007/s00792-005-0476-x
Li B, Liu J, Zhou S, Fu L, Yao P, Chen L, Yang Z, Wang X, Zhang XH (2020) Vertical variation in Vibrio community composition in Sansha Yongle Blue Hole and its ability to degrade macromolecules. Mar Life Sci Technol 2:60–72. https://doi.org/10.1007/s42995-019-00003-4
Lim B, Beyhan S, Yildiz FH (2007) Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL domain protein, in Vibrio cholerae. J Bacteriol 189:717–729. https://doi.org/10.1128/JB.00834-06
Lin IT, Tulman ER, Geary SJ, Zhou X (2021) A gatekeeper protein contributes to T3SS2 function via interaction with an ATPase in Vibrio parahaemolyticus. Microbiol Res 252:126857. https://doi.org/10.1016/j.micres.2021.126857
Liu J, Zheng Y, Lin H, Wang X, Li M, Liu Y, Yu M, Zhao M, Pedentchouk N, Lea-Smith DJ, Todd JD, Magill CR, Zhang WJ, Zhou S, Song D, Zhong H, Xin Y, Yu M, Tian J, Zhang XH (2019) Proliferation of hydrocarbon-degrading microbes at the bottom of the Mariana Trench. Microbiome 7:1–13. https://doi.org/10.1186/s40168-019-0652-3
Liu M, Zhu X, Zhang C, Zhao Z (2021) LuxQ-LuxU-LuxO pathway regulates biofilm formation by Vibrio parahaemolyticus. Microbiol Res 250:126791. https://doi.org/10.1016/j.micres.2021.126791
Lundsten L, Schlining KL, Frasier K, Johnson SB, Kuhnz LA, Harvey JBJ, Clague G, Vrijenhoek RC (2010) Time-series analysis of six whale-fall communities in Monterey Canyon, California, USA. Deep-Sea Res PT I 57:1573–1584. https://doi.org/10.1016/j.dsr.2010.09.003
Lv T, Dai F, Zhuang Q, Zhao X, Shao Y, Guo M, Lv Z, Li C, Zhang W (2020) Outer membrane protein OmpU is related to iron balance in Vibrio alginolyticus. Microbiol Res 230:126350. https://doi.org/10.1016/j.micres.2019.126350
Missiakas D, Raina S (1998) The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol 28:1059–1066. https://doi.org/10.1046/j.1365-2958.1998.00865.x
Missiakas D, Betton JM, Raina S (1996) New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol 21:871–884. https://doi.org/10.1046/j.1365-2958.1996.561412.x
Nakano M, Takahashi A, Su Z, Harada N, Mawatari K, Nakaya Y (2008) Hfq regulates the expression of the thermostable direct hemolysin gene in Vibrio parahaemolyticus. BMC Microbiol 8:155. https://doi.org/10.1186/1471-2180-8-155
Newell CR, Pilskaln CH, Robinson SM, MacDonald BA (2005) The contribution of marine snow to the particle food supply of the benthic suspension feeder, Mytilus edulis. J Exp Mar Biol Ecol 321:109–124. https://doi.org/10.1016/j.jembe.2005.01.006
Oger PM, Jebbar M (2010) The many ways of coping with pressure. Res Microbiol 161:799–809. https://doi.org/10.1016/j.resmic.2010.09.017
Okuno D, Iino R, Noji H (2011) Rotation and structure of F0F1-ATP synthase. J Biochem 149:655–664. https://doi.org/10.1093/jb/mvr049
Paterson GK, Northen H, Cone DB, Willers C, Peters SE, Maskell DJ (2009) Deletion of tolA in Salmonella Typhimurium generates an attenuated strain with vaccine potential. Microbiology 155:220–228. https://doi.org/10.1099/mic.0.021576-0
Phuvasate S, Su YC (2015) Efficacy of low-temperature high hydrostatic pressure processing in inactivating Vibrio parahaemolyticus in culture suspension and oyster homogenate. Int J Food Microbiol 196:11–15. https://doi.org/10.1016/j.ijfoodmicro.2014.11.018
Phuvasate S, Su YC (2015) Alteration of structure and cellular protein profiles of Vibrio parahaemolyticus cells by high pressure treatment. Food Control 50:831–837. https://doi.org/10.1016/j.foodcont.2014.10.037
Raghunath P (2015) Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front Microbiol 5:805. https://doi.org/10.3389/fmicb.2014.00805
Reyes C, Hodgskiss LH, Kerou M, Pribasnig T, Abby SS, Bayer B, Kraemer SM, Schleper C (2020) Genome wide transcriptomic analysis of the soil ammonia oxidizing archaeon Nitrososphaera viennensis upon exposure to copper limitation. ISME J 14:2659–2674. https://doi.org/10.1038/s41396-020-0715-2
Rice SA, Oliver JD (1992) Starvation response of the marine barophile CNPT-3. Appl Environ Microbiol 58:2432–2437. https://doi.org/10.1128/aem.58.8.2432-2437.1992
Ruhal R, Kataria R (2021) Biofilm patterns in gram-positive and gram-negative bacteria. Microbiol Res 251:126829. https://doi.org/10.1016/j.micres.2021.126829
Sato M, Machida K, Arikado E, Saito H, Kakegawa T, Kobayashi H (2000) Expression of outer membrane proteins in Escherichia coli growing at acid pH. Appl Environ Microb 66:943–947. https://doi.org/10.1128/AEM.66.3.943-947.2000
Schwarz JR, Colwell RR (1974) Effect of hydrostatic pressure on growth and viability of Vibrio parahaemolyticus. Appl Environ Microbiol 28:977–981. https://doi.org/10.1128/am.28.6.977-981.1974
Scoma A, Heyer R, Rifai R, Dandyk C, Marshall I, Kerckhof FM, Marietou A, Boshker HTS, Meysman FJR, Malmos KG, Vosegaard T, Vermeir P, Banat IM, Benndorf D, Boon N (2019) Reduced TCA cycle rates at high hydrostatic pressure hinder hydrocarbon degradation and obligate oil degraders in natural, deep-sea microbial communities. ISME J 13:1004–1018. https://doi.org/10.1038/s41396-018-0324-5
Su YC, Liu C (2007) Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol 24:549–558. https://doi.org/10.1016/j.fm.2007.01.005
Sun X, Liu T, Peng X, Chen L (2014) Insights into Vibrio parahaemolyticus CHN25 response to artificial gastric fluid stress by transcriptomic analysis. Int J Mol Sci 15:22539–22562. https://doi.org/10.3390/ijms151222539
Thatje S, Hillenbrand CD, Larter R (2005) On the origin of Antarctic marine benthic community structure. Trends Ecol Evol 20:534–540. https://doi.org/10.1016/j.tree.2005.07.010
Thompson FL, Iida T, Swings J (2004) Biodiversity of vibrios. Microbiol Mol Biol R 68:403–431. https://doi.org/10.1128/MMBR.68.3.403-431.2004
Vezzi A, Campanaro S, D’angelo M, Simonatol F, Vitulo N, Lauro FM, Cestarog A, Malacridab G, Simionatin B, Cannata N, Romualdi C, Bartlett DH, Valle G (2005) Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459–1461. https://doi.org/10.1126/science.1103341
Wagner GP (2012) Measurement of mRNA abundance using RNA-seq data RPKM measure is inconsistent among samples. Theory Biosci 131:281–285. https://doi.org/10.1007/s12064-012-0162-3
Wang Y, Fan L, Tuyishime P, Liu J, Zhang K, Gao N, Zhang Z, Ni X, Feng J, Yuan Q, Ma H, Zheng P, Sun J, Ma Y (2020) Adaptive laboratory evolution enhances methanol tolerance and conversion in engineered Corynebacterium glutamicum. Commun Biol 3:1–15. https://doi.org/10.1038/s42003-020-0954-9
Wang H, Zhang Y, Bartlett DH, Xiao X (2021) Transcriptomic analysis reveals common adaptation mechanisms under different stresses for moderately piezophilic bacteria. Microb Ecol 81:617–629. https://doi.org/10.1007/s00248-020-01609-3
Watnick PI, Lauriano CM, Klose KE, Croal L, Kolter R (2001) The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol Microbiol 39:223–235. https://doi.org/10.1046/j.1365-2958.2001.02195.x
Welch TJ, Farewell A, Neidhardt FC, Bartlett DH (1993) Stress response of Escherichia coli to elevated hydrostatic pressure. J Bacteriol 175:7170–7177. https://doi.org/10.1128/jb.175.22.7170-7177.1993
Xu C, Wang S, Ren H, Lin X, Wu L, Peng X (2005) Proteomic analysis on the expression of outer membrane proteins of Vibrio alginolyticus at different sodium concentrations. Proteomics 12:3142–3152. https://doi.org/10.1002/pmic.200401128
Yin WL, Zhang N, Xu H, Gong XX, Long H, Ren W, Zhang X, Cai XN, Huang AY, Xie ZY (2021) Stress adaptation and virulence in Vibrio alginolyticus is mediated by two (p) ppGpp synthetase genes, relA and spoT. Microbiol Res 253:126883. https://doi.org/10.1016/j.micres.2021.126883
Zhai Z, Yang Y, Wang H, Wang G, Ren F, Li Z, Hao Y (2020) Global transcriptomic analysis of Lactobacillus plantarum CAUH2 in response to hydrogen peroxide stress. Food Microbiol 87:103389. https://doi.org/10.1016/j.fm.2019.103389
Zhang Y, Li X, Bartlett DH, Xiao X (2015) Current developments in marine microbiology: high-pressure biotechnology and the genetic engineering of piezophiles. Curr Opin Biotech 33:157–164. https://doi.org/10.1016/j.copbio.2015.02.013
Zhang Y, Liu H, Gu D, Lu X, Zhou X, Xia X (2020) Transcriptomic analysis of PhoR reveals its role in regulation of swarming motility and T3SS expression in Vibrio parahaemolyticus. Microbiol Res 235:126448. https://doi.org/10.1016/j.micres.2020.126448
Zhang N, Zhang S, Ren W, Gong X, Long H, Zhang X, Cai X, Huang A, Xie Z (2021) Roles of rpoN in biofilm formation of Vibrio alginolyticus HN08155 at different cell densities. Microbiol Res 247:126728. https://doi.org/10.1016/j.micres.2021.126728
Zhou M, Huang Y, Zhang Y, Wang Q, Ma Y, Shao S (2022) Roles of virulence regulator ToxR in viable but non-culturable formation by controlling reactive oxygen species resistance in pathogen Vibrio alginolyticus. Microbiol Res 254:126900. https://doi.org/10.1016/j.micres.2021.126900
Zobell CE, Cobet AB (1964) Filament formation by Escherichia coli at increased hydrostatic pressures. J Bacteriol 87:710–719. https://doi.org/10.1128/jb.87.3.710-719.1964
Funding
This work was supported by the National Key Research and Development (R&D) Program of China (2017YFC1600703).
Author information
Authors and Affiliations
Contributions
SL: writing—original draft, formal analysis, resources, writing—review and editing. TZ: conceptualization, methodology, validation. ZL: data curation, resources. JW: data curation, investigation, visualization. CZ: language editing. QK: language editing, visualization. XF: supervision, conceptualization, writing—review and editing. HM: supervision, conceptualization, writing—review and editing, funding acquisition.
Corresponding authors
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liang, S., Zhang, T., Liu, Z. et al. Response mechanism of Vibrio parahaemolyticus at high pressure revealed by transcriptomic analysis. Appl Microbiol Biotechnol 106, 5615–5628 (2022). https://doi.org/10.1007/s00253-022-12082-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-12082-y