Abstract
Context
Understanding genetic structure at multiple spatial scales and identifying drivers of genetic isolation are important for developing comprehensive conservation plans including for grassland conservation efforts. However, few studies account for multiple genetic isolation processes nor partition genetic variance among these processes.
Objectives
We assess key processes that can create spatial genetic patterns including isolation by barrier (IBB), isolation by distance (IBD), and isolation by environment (IBE) for a widespread pocket gopher species (Geomys bursarius) and a spatially restricted subspecies (Geomys bursarius illinoensis). We further partition genetic variation to each isolating effect and identify genetic variation that was shared between processes.
Methods
We used seven microsatellites to determine spatial genetic clustering and identify environmental factors impacting genetic similarities. Then, we used redundancy analysis to partition variance explained by IBB, IBD, and IBE.
Results
Major rivers including the Mississippi River acted as barriers and explained the most genetic variation across the species. In contrast, IBD explained the most genetic variation for G. b. illinoensis. Gophers had genetic associations to soil sand percent and soil color, but IBE uniquely explained a small amount of genetic structure for G. bursarius, with additional variation shared with other isolating processes.
Conclusions
Gopher genetic structure resulted from barriers, distance, and environmental factors at the species range as well as for a subspecies’ region, but the relative amount of genetic variance assigned to unique isolating processes differed between scales. Delineation of conservation units should consider major rivers as natural boundaries, and finer-scale management should identify and protect areas close to source populations with similar soil friability. Our study exemplifies how analyzing gene flow at rangewide and regional scales can aid managers in developing localized strategies that fit within broader conservation units.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetic diversity is a central component of biodiversity (DeWoody et al. 2021). To protect biodiversity, therefore, we must understand how different processes structure genetic variation across temporal and spatial scales (Hewitt 2004; Cushman and Landguth 2010; Lucati et al. 2020). Key processes include isolation by barrier (IBB) in which impermeable landscape elements cause genetic isolation by restricting gene flow (Manel et al. 2003; Holderegger and Wagner 2008; Manel and Holderegger 2013), isolation by distance (IBD) that produces continuous genetic differentiation due to spatially limited dispersal (Wright 1943; Jenkins et al. 2010; van Strien et al. 2015), and isolation by environment (IBE) in which heterogeneous environments may further impact gene flow beyond IBD due to local adaptation or natal-site selection (Shafer and Wolf 2013; Sexton et al. 2014; Wang and Bradburd 2014).
These three processes may all contribute to spatial genetic variation, however, few studies have quantified their relative effects. For instance, Muñoz-Valencia et al. (2023) found that IBD and IBE effects from precipitation and temperature explained the most genetic variance of leaf-cutter ants (Atta cephalotes), but additional genetic variance was explained by the Andes Mountains acting as a barrier. Likewise, genetic variance of American badgers (Taxidea taxus) was explained by geographic distance, the Wisconsin River as a barrier, and agricultural land cover (Kierepka and Latch 2016a). Through partitioning genetic variation to these distinct processes, we can gain a holistic understanding of spatial genetic structure (Weber et al. 2016; Priadka et al. 2019; Muñoz-Valencia et al. 2023).
IBB patterns commonly arise from mountain ranges (Muñoz-Valencia et al. 2023), rivers (Pfau et al. 2001; Musher et al. 2022), or other impassible landscape structures. However, semi-permeable barriers may exist, including from anthropogenic causes (e.g., roads), where gene flow may be reduced but not completely disrupted (Esperandio et al. 2019; Lecis et al. 2022). IBD results from short dispersal distances across geographic space. Genetic similarities may be greater than expected under IBD, however, because of rare long-distance dispersal that maintains infrequent gene flow (Centeno-Cuadros et al. 2011; Alexander et al. 2019). Whereas IBB and IBD arise from dispersal processes, gene flow can also be driven by characteristics within a species’ home range, leading to genetic patterns in neutral markers. IBE arises when local adaptation or natal habitat selection creates genetic structure due to associations with specific environmental conditions rather than solely spatial processes (Shafer and Wolf 2013; Orsini et al. 2013; Wang and Bradburd 2014). A combination of these processes affecting gene flow may be intertwined in species that span large riverways or other barriers, persist across a large geographic range, or exhibit strong environmental associations.
Furthermore, these genetic isolating effects may be spatially dependent with different processes dominating rangewide versus within a specific region (Anderson et al. 2010; Keller et al. 2013). For instance, IBB is expected to be most consequential at larger scales that may include multiple substantial barriers to movement. IBD and IBE may be strongest at smaller scales reflecting dispersal and habitat selection occurring within regions defined by barriers. IBD is more pronounced for species with low dispersal (Orsini et al. 2013), but may exhibit a threshold pattern, where eventually geographic distance does not contribute to added genetic distance (van Strien et al. 2015). Similarly, IBE may vary across spatial scales either due to environmental spatial autocorrelation (Shafer and Wolf 2013), or dispersal swamping out local adaptation (Richardson et al. 2014). Determining the relative strengths and scales of the isolating processes can guide efforts to maintain or restore suitable habitat and landscape connectivity.
Multi-scale landscape genetic studies can clarify management units and identify the spatial extent that matches conservation goals (Keller et al. 2015). By retaining large-scale management considerations, restoration or habitat management at small scales can be leveraged for broad-scale planning (Augustine et al. 2021, Gilby et al. 2021). This perspective is important for managing prairie ecosystems as large-scale, connected grasslands are needed for both migratory species and sedentary species with metapopulation dynamics (Augustine et al. 2021). However, grasslands often are small and fragmented patches with highly altered vegetation communities, thus identifying how local management practices fits within a rangewide context is necessary to promote connectivity (Augustine et al. 2021; Warner 1994).
The plains pocket gopher (Geomys bursarius) is a grassland species that provides an excellent opportunity to examine multiple drivers of gene flow across spatial scales because it is a subterranean species that may have strong, hierarchical genetic structuring (e.g., Mapelli et al. 2020). Geomys bursarius ranges across the Great Plains from Texas to southern Canada, with only G. b. illinoensis and G. b. wisconsinensis occurring east of the Mississippi River (Connior 2011), likely following post-glacial range expansion similar to many other taxa (Smith 1957). Rivers and other water bodies are likely effective barriers that prevent gene flow for many subterranean rodents (Visser et al. 2018; Mapelli et al. 2020; Austrich et al. 2020). G. bursarius cannot swim well (Komarek and Spencer 1931; Kennerly 1963), and the Mississippi River aligns with a subspecies boundary for G. b. illinoensis (Connior 2011). Furthermore, most subterranean rodents have limited gene flow, even over short distances, and develop IBD over time (Mapelli et al. 2012; Fasanella et al. 2013; Gómez Fernández et al. 2016; Visser et al. 2018). Geomys bursarius also exhibits strong soil associations (Reichman and Seabloom 2002), and anthropogenic land use prevents gophers from persisting in historically occupied soils (Alexander et al. 2022). Understanding these isolating effects may help to designate management units and ensure conservation efforts are enacted close enough to an already established population.
Burrowing rodents often display local adaptation to soil structure, potentially generating IBE patterns (Barbosa et al. 2021). In fact, soil properties act as distribution boundaries that delineate gopher species (Hoffman et al. 2007; Hoffman and Choate 2008). Gophers may exhibit IBE patterns based on energetic costs related to soil friability (Vleck 1979, 1981) and selection of familiar soil types. Also, gophers exhibit pelage matching to soil color (Hendricksen 1972; Krupa and Geluso 2000; Rios and Álvarez-Castañeda 2012), which decreases predation risk even for subterranean rodents (Krupa and Geluso 2000; Rios and Álvarez-Castañeda 2012; Singaravelan et al. 2013). Although gopher pelage color has not been linked to a single locus (Wlasiuk and Nachman 2007), neutral loci may identify how soil color influences gene flow due to presumed fitness costs.
There are two possible reasons for pelage color matching soil color. First, gophers occasionally disperse above ground (Warren et al. 2017; Pynne et al. 2019), and matching surface soil color would increase crypsis during dispersal events. Second, crypsis during burrow construction may impact fitness. How mound creation correlates to time above ground is unclear, but gophers may create three mounds per day with ~ 60% of excavated soil deposited on the surface (Andersen 1987). Gopher foraging tunnels are close to the surface, whereas nest chambers may be deeper (~ 50 to 100 cm below the surface; Wilkins and Roberts 2007). Crypsis during soil removal from burrows may drive color-matching adaptation. However, genetic patterns resulting from fitness benefits of crypsis in gophers has not been assessed. Identifying genetic associations with soil properties is also important for conservation as gophers may be excluded from historically occupied soils due to land-use intensification (Alexander et al. 2022).
Because IBB, IBD, and IBE may all contribute to genetic structure, we examine the relative effects of these isolating processes on plains pocket gophers. Also, because genetic differentiation is scale dependent, we explore isolating effects both for G. bursarius across the geographic range and for a subspecies, G. b. illinoensis, within a region including Illinois and Indiana. Geomys bursarius illinoensis should be considered an Evolutionary Significant Unit (Alexander 2023), and it occurs in a region historically dominated by tallgrass prairie and oak savannah but now dominated by agriculture (Augustine et al. 2021; Alexander et al. 2022). For G. bursarius, we predict that IBB effects will occur due to the Mississippi River and other major waterways, and that there will be strong IBD due to limited dispersal. We also hypothesize there will be IBE from soil sand percentage affecting friability and from soil surface color affecting crypsis during above ground dispersal and creating mounds. For G. b. illinoensis, we predict no IBB effects due to no large riverways within the subspecies range, but we expect IBD from dispersal limitations and colonization history, and IBE due to similar processes for the species across the range.
Methods
Tissue samples and DNA extraction
We collected toe samples from 241 museum specimens of G. bursarius from the Illinois Natural History Survey including 8 subspecies from across the range, only omitting G. b. ozarkensis (Table S1). All museum specimens were collected between 1921 and 1985 and included subspecies identification except for samples from Wisconsin (n = 30), which we classified as G. b. wisconsinensis based on the range map (Connior 2011). We also collected tissue samples from 27 live-trapped G. bursarius illinoensis (UIUC IACUC #17190) in 2018–2019, following appropriate guidelines (Sikes and The Animal Use and Care Committee of the American Society of Mammalogists 2016). To improve amplification, we used a 1× Sodium Choride-Tris–EDTA buffer to increase solubility of DNA and lysed the sample in an incubator with a rotisserie at 56 ˚C. We used QIAquick spin columns to capture DNA. We successfully sampled and amplified a total of 267 G. bursarius, 170 of which were G. b. illinoensis.
Microsatellite amplification and verification
To identify genetic structure, neutral genetic markers (e.g., microsatellites) are commonly used as they are not under selection and have a high mutation rate (Epps and Keyghobadi 2015). Although genomic or other markers (e.g., SNPs) are gaining use, microsatellites can still identify similar environmental-genetic associations and underlying processes (Hauser et al. 2021; Skey et al. 2023). We attempted amplification of 12 microsatellites (GBR06, GBR09, GBR10, GBR15, GBR25, GBR26, GBR27, GBR33, GBR36, TM1, TM6, and TM7; Steinberg 1999; Welborn et al. 2012). We optimized polymerase chain reactions by trying different starting conditions, and fragment analysis was done on an ABI Prism 370xl Analyzer. Alleles were scored manually using Geneious v.11.1.5 (https://www.geneious.com).
To assess population heterozygosity statistics and determine that genetic markers were not closely associated with one another, we initially assigned individuals to populations a priori based on spatial proximity, using major rivers in the United States as population boundaries (Supplemental 1; Esri et al. 2010). We assessed Hardy–Weinberg Equilibrium (HWE), linkage disequilibrium (LD), and null alleles using R code adapted from Wagner (2022). To test for HWE, we used a conservative ɑ = 0.05 with a chi-square test using the function “hw.test” in the R package pegas (Paradis 2010). If a locus was consistently out of HWE across populations, the locus was dropped. We tested LD via an index association analysis, using the “ia” function (Brown et al. 1980) and loci association using “pair.ia” (Agapow and Burt 2001) in poppr (Kamvar et al. 2014) with a permutation of 999. If the measure of correlation (̄r̄d) was ≤ 0.3 from pairwise comparisons between loci, the loci were retained as this would approximate a 10% linear correlation threshold (Wagner 2022). We estimated null alleles in PopGenReport (Adamack and Gruber 2014) using the “null.all” function, implementing the Brookfield (1996) method that effectively estimates null alleles at loci that could falsely appear homozygous. If there is spatial structuring and fixed alleles from Wahlund or other isolating effects, there may be an overestimation of null alleles, as null allele estimates assume HWE (Dabrowski et al. 2014). Thus, we assessed null alleles rangewide and for each region independently. We also estimated observed and expected heterozygosity and calculated rarefied allelic richness in PopGenReport (Adamack and Gruber 2014) and fixation index (FST) and inbreeding coefficient (FIS) in hierfstat (Goudet 2005). Two regions had low sample sizes (n ≤ 8). Given that this could impact our region-specific estimates of heterozygosity, rarefied allelic richness, and Fst, we estimated gene flow and population structure relying on individual-based analyses.
Isolation by barriers
To determine whether genetic barriers exist across the geographic range of G. bursarius, we used STRUCTURE v. 2.3.4, which identifies genetic clusters through maximizing HWE (Pritchard et al. 2000; Falush et al. 2003). STRUCTURE performs well at identifying the highest level of discrete genetic structures (Evanno et al. 2005; Chen et al. 2007), and it can also identify hierarchical genetic structures (Warnock et al. 2010).
We ran STRUCTURE with an admixture model without location priors (Hubisz et al. 2009). We tested 1–10 populations (K) with 5 trials at each value of K with 300,000 iterations and a 100,000-iteration burn-in. To determine the number of genetic clusters, we used the Evanno method that relies on the change in likelihood (Evanno et al. 2005) using STRUCTURE HARVESTER (Earl and vonHoldt 2012). Because STRUCTURE’s log probability often overestimates K, the Evanno method identifies the highest hierarchical genetic clustering based on the log probability’s rate of change between successive K values (Evanno et al. 2005). We then visually inspected assignment of individuals to each cluster using CLUMPAK (Kopelman et al. 2015) using a 0.5 population assignment threshold (Kierepka and Latch 2016a). Because STRUCTURE resolves the highest level of genetic structure, we re-ran STRUCTURE on each identified cluster to determine if there was hierarchical genetic structure until no spatial pattern was evident (Vähä et al. 2007; Warnock et al. 2010). Then, we re-ran STRUCTURE just on G. b. illinoensis samples to determine genetic clusters within a single subspecies across a smaller spatial scale.
Uneven sampling can impact STRUCTURE clustering (Puechmaille 2016), so we thinned the G. b. illinoensis samples by 10 km (n = 40), keeping only one sample if multiple individuals were sampled within that distance, and re-ran STRUCTURE. Given the smaller sample size of the other seven subspecies (n = 97 total), we maintained all samples that were not G. b. illinoensis regardless of nearest-neighbor distance. G. b. illinoensis still had the highest sample size but was comparable to other subspecies (e.g., n = 32 for G. b. wisconsinensis). GBR06 amplification failed mostly in northern samples, so we ran STRUCTURE using the thinned sampling of G. b. illinoensis and all other Geomys bursarius samples excluding GBR06 to ensure amplification failure did not bias clustering.
To further understand how rivers may act as barriers for gophers, we calculated FST (Weir and Cokerham 1984) between a priori regions (see Section “Microsatellite amplification and verification”). Whereas STRUCTURE can create clusters based on genetics with landscape features being identified post hoc¸ estimating FST between regions allows for testing of our IBB hypothesis directly. For G. bursarius rangewide, we assigned individuals to populations with major rivers acting as population boundaries (Supplemental 1, Esri et al. 2010). For G. b. illinoensis, we assigned individuals to populations with rivers that form major watersheds (Illinois State Water Survey 2011) serving as population boundaries. To calculate FST, we used “pairwise.WCfst” and estimated bootstrap values with “boot.ppfst” and 1000 bootstraps in the R package hierfstat (Goudet 2005).
Isolation by distance
To detect genetic spatial autocorrelation for G. bursarius (rangewide) and for G. b. illinoensis (within Illinois and Indiana), we used Moran Eigenvector Maps (MEMs), which rely on regression of genetic distances in a multivariate framework (Galpern et al. 2014). MEMs identify genetic clines across geographic distances, making them a complementary approach to STRUCTURE (Galpern et al. 2014; Priadka et al. 2019). We calculated proportion of shared alleles (DPS; Bowcock et al. 1994) to estimate genetic dissimilarity between individuals (Shirk et al. 2017) using the “codomToPropShared,” function and then calculated and visualized Moran eigenvectors using the “mgQuick” and “mgMap” functions in the R package memgene (Galpern et al. 2014), mapping the first two axes. We also ran a leave-one-out sensitivity analysis of MEMs, iteratively omitting one locus to determine potential effects of null alleles.
Isolation by environment
We tested if gene flow was associated with soil conditions using redundancy analysis (RDA) with a Principal Components Analysis (PCA) of allele frequencies as a dependent matrix (Muñoz-Valencia et al. 2023). We conducted the analysis for G. bursarius and at the regional scale for G. b. illinoensis. In our global model, we included sand percent (Soil Survey Staff 2018) as a metric of soil friability resulting from glacial processes (Dobos et al. 2023), soil color at 5 cm and 75 cm depths (Soil Survey Staff 2022) to assess genetic signatures of pelage-soil color matching, and geographic distance to account for IBD genetic patterns. Although multiple soil properties may impact friability (e.g., percent clay, particle size), these parameters were all correlated with soil sand percent. Thus, soil sand percent should be considered as a proxy for multiple contributors to soil friability. Land cover is an increasingly important driver for G. bursarius (Alexander et al. 2022), however, given the dynamic nature of land cover and the temporal breadth of our genetic samples, we retained only predictors that should be largely stable over the past 100 years. To determine if there were temporal effects on genetic similarity, we included the year of sample collection in the model.
We included soil color at two depths as color matching may occur for above ground dispersal (5 cm) or for excavation of deeper soils to create nest or other chambers (75 cm) and depositing those soils on the surface. We converted soil color maps to RGB color maps in ArcMap 10.8.1 using the “copy raster” function, in which a unique raster was generated for red, green, and blue pixel values. We then calculated Euclidean distances in three-dimensional space between the raster values at gopher locations for soil color at 5 cm and 75 cm depths to quantify soil color similarity. To convert distance matrices to linear vectors, we used principal coordinates of neighborhood matrices (PCNM; Borcard and Legendre 2002) for geographic distance and soil color distances using the “pcnm” function in the R package vegan (Oksanen et al. 2022). All continuous variables were scaled by centering means to 0 and dividing by the standard deviation. We used “dudi.pca” from the ade4 R package (Dray and Dufour 2007) and retained two principal components (PCs) of the allele frequencies PCA. We then performed RDAs using the “rda” function to create a global and a null model and used “ordistep” in vegan to select the top models for G. bursarius and then for G. b. illinoensis. We assessed model fit using an Analysis of Variance using the “anova.cca” function and estimated the adjusted R2 using the “RsquareAdj” function in vegan. We assessed variation inflation factors (VIFs) using the “vif.cca” function in vegan. We also estimated IBE following the above methods omitting each locus iteratively in a leave-one-out sensitivity analysis to determine potential impacts of null alleles.
Variation partitioning
To determine the relative effects of IBB, IBD, and IBE on gopher genetic structure, we conducted variation partitioning of the two PCs for G. bursarius and for G. b. illinoensis (see ‘Isolation by Environment’). We ran “rda,” “vif.cca,” and “varpart” in vegan (see Section “Isolation by environment”), separating out a priori regions for IBB (see Section “Microsatellite amplification and verification”), PCNM of distance for IBD, and then PCNM of soil colors (5 and 75 cm) and sand percent for IBE. We also included the year of sample collection to account for genetic similarity based on time. We estimated variation partitioning omitting each locus iteratively in a leave-one-out sensitivity analysis to determine potential impacts of null alleles.
Results
Microsatellite amplification and verification
Four microsatellites (GBR15, GBR26, GBR33, and TM7) either had amplification failure or ambiguous genotypes, and one microsatellite (GBR36) was monomorphic. These five microsatellites were excluded from analyses, leaving a total of seven microsatellites. Other studies have effectively identified genetic structure with a similar number of markers (e.g., Cosentino et al. 2015; McCluskey et al. 2022). We also identified substantial genetic structuring (see below), but we acknowledge that additional fine-scale structure may have gone undetected.
All retained loci were out of HWE globally; however, no locus was consistently out of HWE across all a priori populations determined by major rivers indicating Hardy–Weinberg disequilibrium was a result of spatial isolation and drift rather than other mechanisms. The population of G. b. illinoensis deviated from HWE at all loci, but this is likely due to increased sample size compared to the other regions. The Wisconsin samples only had one individual that amplified at GBR06, so GBR06 was omitted from the Wisconsin null allele analysis. One locus, TM6, had null alleles in regions with higher sample sizes, so we estimated observed heterozygosity, expected heterozygosity, and rarefied allelic richness excluding TM6 as well (Supplemental 2). Null allele frequencies ranged from 0.1 to 0.34 (Supplemental 2). Deficiencies of heterozygotes can increase the signal null alleles (Dabrowski et al. 2014; Meeûs 2018), and identification of null alleles at the rangewide scale is likely due in part to population structure (i.e., Wahlund Effects). Within regions, null alleles were generally lower, with estimates often not distinguishable from 0 (Supplemental 2). Given the low dispersal distances of G. bursarius and likely Wahlund effects, we kept the remaining 7 microsatellites in the analyses. The overall loci index association was 0.22 and an ̄r̄d = 0.038. However, because all pairwise ̄r̄d were ≤ 0.3, we assumed no linkage between loci. Sample sizes from apriori populations ranged from 4 to 170 (mean = 38, SE = 22; Supplemental 2). Observed heterozygosity was lower than expected heterozygosity across all loci globally (Ho = 0.477, He = 0.702, FST = 0.167, FIS = 0.320; Supplemental 2). Rarefied allelic richness ranged from 2.14 to 2.88 (Supplemental 2).
Isolation by barrier
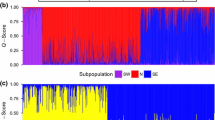
For G. bursarius, we identified 2 populations across the range with the STRUCTURE analysis. At this highest level, G. b. illinoensis clustered separately from the rest of G. bursarius with the separation mostly aligning with the Mississippi River (Fig. 1a). We then observed genetic clustering west of the Mississippi River (n = 103) with two populations identified mostly along subspecies and latitudinal gradients (nK1 = 57, nK2 = 47), generally separated by the Missouri River (Fig. 1b). For the G. b. illinoensis population east of the Mississippi River (n = 164), two subpopulations were identified (nK1 = 101, nK2 = 63) but admixture in the individual assignment plots revealed a clinal structure rather than discrete populations (Fig. 1c). These two subpopulations within G. b. illinoensis are likely spurious clusters due to STRUCTURE’s algorithm and more likely represent an IBD pattern.
Individual population assignments with admixture from STRUCTURE (left), and number of populations estimated using the Evanno method (right) without spatial thinning for a all samples of Geomys bursarius (n = 267), b with further structuring for the western (n = 103) and eastern (n = 164) populations, and c only G. b. illinoensis (n = 170). Individual assignments viewed on the base World Topography Map (ESRI et al. 2017)
Using a thinned sample of G. b. illinoensis but with all other samples included, we still identified two populations, but the clusters resolved around a south-north barrier. The structure included populations north (n = 52) and south (n = 85) of the Missouri and Illinois Rivers, with G. b. illinoensis clustering with the southern samples (Fig. 2a). Despite two populations having the strongest support, three populations were also well supported. We visually inspected individual assignments with K = 3, which identified G. b. illinoensis as a third cluster (Supplemental 3).
Population assignments from STRUCTURE (left) and number of populations estimated using the Evanno method (right) with (a) spatial thinning of Geomys bursarius illinoensis (n = 137), and b samples from the resulting substructures in the northern population (n = 52) and the southern population (n = 85) identified from the initial clustering with K = 3 in the north and K = 2 in the south. There is close support for K = 3 for the initial clustering (a), and close support for K = 2 for the northern population (b). Individual assignments viewed on the base World Topography Map (ESRI et al. 2017) with major rivers in the US (Esri 2010)
For the two clusters identified with the thinned sampling method, no further spatial structure resolved in the northern population, indicating that the seven microsatellites used here could not separate the subspecies G. b. majusculus, G. b. wisconsinensis, and G. b. bursarius. For the southern cluster, there was added structure with G. b. illinoensis clustering separately, and the Mississippi River acting as a boundary (Fig. 2b). When G. b. illinoensis was analyzed alone, we identified 2 populations (Fig. 1c). However, individuals showed a south-north gradient of admixture indicating that IBD is more likely than IBB for G. b. illinoensis.
For G. bursarius, FST values ranged from 0.048 to 0.320 based on a priori defined regions divided by major rivers (Supplemental 2). The populations divided by the Canadian River in the southern region had relatively low FST (0.049), whereas populations divided by the Arkansas, Missouri, Mississippi, and Illinois Rivers all had pairwise FST > 0.1. FST for only G. b. illinoensis ranged from 0.035 to 0.145 based on region, with only the populations divided by the Canadian River having an Fst of 0.049 (Supplemental 2). The Canadian River divides the Kiowa and Red Hills regions, which both had a low sample size, and further sampling may better identify genetic structure created by the river.
Isolation by distance
For G. bursarius, geographic distance was correlated with genetic distance (r2 = 0.34) based on the MEM. The MEM identified structuring along the first axis similar to STRUCTURE, with two populations divided by the Mississippi River (proportion of variation = 0.41). The second axis revealed a south-north clinal genetic structure (proportion of variation = 0.27; Fig. 3a). The leave-one-out sensitivity analysis displayed the same pattern (Supplemental 4), so only the analysis retaining all loci is included here.
For G. b. illinoensis, geographic distance also was correlated with genetic distance (r2 = 0.17). Gophers in southwest Illinois were genetically similar to gophers in the northeast along the first axis (proportion of variation = 0.46). However, gophers in southwest Illinois were genetically similar to gophers in western Illinois along the second axis (proportion of variation = 0.26; Fig. 3b).
Isolation by environment
For the IBE models, all VIFs were < 2 indicating no multicollinearity and the first two principal components explained genetic variance well (Supplemental 5). For G. bursarius, the top model included soil color at 5 cm (p < 0.001), sand percent (p < 0.001), and geographic distance (p < 0.001) with an adjusted R2 of 0.36 (Table 1). The rangewide leave-one-out analysis mostly included the same parameters, with the model omitting TM6 only including distance and sand percent, and the model omitting GBR25 including distance, soil color at 5 cm, and soil color at 75 cm (Supplemental 6). For G. b. illinoensis, the top model included soil color at 75 cm (p = 0.029), sand percent (p = 0.005), and geographic distance (p < 0.001) with an adjusted R2 of 0.31 (Table 1). For G. b. illinoensis, all of the leave-one-out models included distance, five out of the seven top models included sand color at 75 cm, and four out of the seven top models included sand percent (Supplemental 6).
Variation partitioning
For G. bursarius, IBB explained the most variance independently (adjusted R2 = 0.26), but there was high variance shared by IBB and IBD (adjusted R2 = 0.27; Fig. 4a). Variance partitioned to IBE minimally explained genetic patterns independently (adjusted R2 = 0.01). However, additional variance was shared by IBB and IBE (adjusted R2 = 0.04) and by IBB, IBD, and IBE (adjusted R2 = 0.03). IBD did not explain any genetic variance independently rangewide, but shared variance with IBB and IBE. Year did not have any variation partitioned independently, but had variance shared by IBB and IBD (adjusted R2 = 0.02). Similar patterns emerged from the leave-one-out sensitivity analysis (Supplemental 6).
Partitioning of genetic variation between isolation by environment (IBE), isolation by barrier (IBB), isolation by distance (IBD), and year. Results are for a Geomys bursarius using 2 principal components (PCs) of genetic variation, and b G. b. illinoensis using 2 PCs of genetic variation. Values < 0 are not shown and values of 0.00 indicate low partitioning values rounded to 0
For G. b. illinoensis, variance shared by IBB and IBD was relatively strong (adjusted R2 = 0.18), but IBD had the most variation partitioned as a single factor (adjusted R2 = 0.06; Fig. 4b). IBE did not explain any genetic variation independently, but IBE shared variance with IBB and IBD (adjusted R2 = 0.04). Year again had minimal variation partitioned to it. Overall, IBB and IBD processes explained the most genetic variation for the gopher subspecies. Similar patterns emerged from the leave-one-out sensitivity analysis (Supplemental 6).
Discussion
By considering the relative effects of isolation by barriers, distance, and environment, we showed that large-scale and regional gene flow for a fossorial rodent were driven by different processes. For the range-wide analysis of G. bursarius, more genetic variation was partitioned uniquely to IBB than to IBD or IBE. In contrast, when analyses were restricted to G. b. illinoensis in Illinois and Indiana, IBD uniquely explained the most genetic variation. However, substantial genetic variation could not be partitioned between IBB and IBD at either scale likely due to both processes creating north–south isolating effects. Isolation by environment minimally explained genetic structure at both scales, although IBE shared variance with IBB and IBD.
Isolation by barriers
Subterranean species, including gophers, are often isolated by major rivers (Connior 2011; Cutrera et al. 2013; Visser et al. 2018). Fossorial rodents have unique proximal limb morphology compared to rodents with other locomotor ecologies (Hedrick et al. 2020), possibly impacting swimming ability. For instance, G. bursarius can swim for ≤ 6.5 min (Hickman 1977). Major rivers acted as barriers for G. bursarius rangewide, with the Mississippi River, Illinois River, and Missouri River aligning closely with genetic separation identified by STRUCTURE. Both the Missouri and Mississippi Rivers have been identified as gene flow barriers for other taxa including badgers (Kierepka and Latch 2016b), northern leopard frogs (Rana pipiens; Waraniak et al. 2019), and Nearctic milksnakes (Lampropeltis triangulum; Burbrink et al. 2022), and the Illinois River is considered the northern range barrier for G. b. illinoensis (Hoffmeister 1989; Alexander et al. 2022). Geomys bursarius illinoensis likely colonized Illinois from individuals coming from southern populations of G. bursarius. After a dispersal event across the Mississippi during the Pleistocene (Smith 1957; Alexander 2023), rivers have remained important barriers. Moreover, the low observed heterozygosity supports a Wahlund effect of gopher populations isolated by barriers (Penney and Zimmerman 1976), and FIS indicated that there is likely further population structure within each region. With higher resolution markers, further genetic structuring may be detected.
Even major riverways, however, may not have consistent barrier effects. Geomys bursarius on the eastern side of the Mississippi River in Wisconsin clustered with gophers to the west of the Mississippi rather than with G. b. illinoensis. However, this pattern is probably due to colonization history. There were likely two instances of G. bursarius crossing the Mississippi River, once from Missouri into Illinois, and a second within Wisconsin (Elrod et al. 2000). The Illinois River then acted as a barrier between G. b. illinoensis and G. b. wisconsinensis. The Mississippi River is also narrower in the north compared to the Missouri-Illinois divide, so colonization events may have been easier there. With smaller rivers there is likely reduced gene flow, but not complete isolation (Roratto et al. 2015; Painter et al. 2022). This pattern is similar to genetic structure of badgers in Wisconsin, with the Mississippi River acting as a barrier broadly (Kierepka and Latch 2016a), yet the smaller Wisconsin River does not (Kierepka and Latch 2016b).
When restricting the analyses to G. b. illinoensis in Illinois and Indiana, STRUCTURE and MEMs did not identify genetic discontinuity based on rivers. Moreover, most genetic variance was not uniquely associated with IBB and instead could not be partitioned between IBB and IBD. Hence, our prediction of IBB effects being scale-dependent was supported.
Isolation by distance
Although river barriers explained broad genetic variation, IBD patterns also emerged from the MEMs and variation partitioning analyses. For G. bursarius, however, genetic variation could not be partitioned between IBD and IBB due to the barriers isolating regions in an almost linear manner from the southwest to the northeast. The inability to partition between geographic distance and barrier effects may be due to northward colonization post glaciation, where colonization occurred in a largely linear manner. On a finer scale for G. b. illinoensis, IBD emerged as a contributor to genetic structure that could be parsed, but there was still considerable variation shared between IBD and IBB.
An interesting pattern emerged from the MEM of G. b. illinoensis in which the southwestern gophers were genetically similar to the northeastern gophers along the 1st axis but displayed similarities with other western individuals along the 2nd axis. This pattern further indicates that genetic structure remains from colonization history in which the G. b. illinoensis population in the southwest crossed the Mississippi River and expanded northeastward along the southern boundary of the Illinois River (Elrod et al. 2000). Although inferring colonization history from seven microsatellites should be done cautiously, this result is supported from phylogenetics indicating post-glaciation range expansion of G. bursarius across the Mississippi River (Smith 1957; Alexander 2023). An IBD pattern for subterranean species is expected as they likely exhibit short-distance dispersal (Welborn and Light 2014; Warren et al. 2017). Gophers disperse short distances above ground (< 800 m), generally with smaller individuals or juveniles dispersing and recruiting within < 50 m or until they encounter suitable habitat (Vaughan 1963; Williams and Cameron 1984; Daly and Patton 1990; Warren et al. 2017).
Isolation by environment
Isolation by environment also contributed to genetic structure of G. bursarius, although minimally compared to other isolating processes. A key concept of IBE is that neutral loci can detect outcomes of processes driven by habitat selection or local adaptation without necessarily identifying the specific process (Sexton et al. 2014; Wang and Bradburd 2014). Although local adaptation may impact neutral processes, genetic drift and isolation drive mostly swamped any adaptive processes that might be identified. Soil dependency varies among Geomys species (Davis et al. 1938; Wilkins and Swearingen 1990; Alexander et al. 2022) and can maintain genetic and morphometric structure (Hendricksen 1972; Sudman et al. 1987; Mauk et al. 1999; Genoways et al. 2008). An interesting phenomenon that may underly the MEM for G. b. illinoensis in Illinois and Indiana is that soil properties vary across the distribution, with western regions having sandier soils and the northeastern and southwestern regions having a higher clay content. G. b. illinoensis has a bimodal selection for sand percent (Alexander et al. 2022), and with sand percent contributing to genetic variation across spatial scales based on IBE models, soil friability may affect genetic structure of gophers, fitting roughly with the MEM analysis.
Although texture and friability affect genetic structure of gophers, soil color also contributed to the IBE models. Pelage color matching soil color occurs across gopher species, likely due to predation risk (Hendricksen 1972; Krupa and Geluso 2000; Rios and Álvarez-Castañeda 2012). For G. bursarius, soil color at 5 cm depth was included in the top model, indicating soil matching affects genetic structure and there is predation pressure during above-ground dispersal for a predominantly subterranean species (Williams and Cameron 1984; Warren et al. 2017; Pynne et al. 2019). On a smaller scale for G. b. illinoensis, soil color at 75 cm depth contributed to genetic structure, but not at 5 cm. However, this outcome is likely due to limited variation of soil color at 5 cm at genetic sample locations for G. b. illinoensis. Soil color is impacted by soil texture, organic matter, minerals, and hydrology (Wascher et al. 1960; Schulze et al. 1993). Further, oxidation may convert blue-gray soil colors to more of an olive-brown (Donald McKay et al. 1986), thus limiting variation in soil color at 5 cm. G. b. illinoensis has experienced a niche reduction and shift in relation to soil sand percent and texture due to agricultural intensification, with a contemporary bimodal response to soil sand percent and a general shift towards sandier soils (Alexander et al. 2022). The impact of surface soil color on gene flow might be more pronounced with more contemporary samples.
Conclusions
Our complementary analyses demonstrated commonalities as well as differences in genetic structure and environmental associations across spatial scales of a species complex. We illustrated hierarchical genetic structures for a fossorial species in which IBB explained most of the genetic variation. However, IBD and IBE also were consequential processes. Major rivers act as barriers; geographic distance creates clinal structure, at least within our focal subspecies, and soil traits promote genetic structure across spatial scales. As gopher relationships with soils have changed over time due to land use (Alexander et al. 2022), understanding potential gene flow reduction associated with loss of habitable soils can inform management decisions.
Whereas management of G. bursarius rangewide should focus on discrete management units bounded by rivers acting as natural barriers, management of G. b. illinoensis should consider ensuring gophers can reach suitable sites and that have similar soil properties as the source population. This allows managers to focus local efforts to bolster connectivity for populations within management units. Specifically, the Conservation Reserve Program is a strong tool to help manage grassland patch connectivity (Augustine et al. 2021), but managers applying these and similar conservation programs also should consider soil friability and distance to source populations when considering site prioritization. As gophers are one of many species that expanded northward post-glaciation, colonizing east of the Mississippi River during the Pleistocene (Smith 1957), partitioning genetic isolating effects can identify conservation priorities for a multitude of species.
More generally, recognizing genetic-environmental associations is increasingly important for conservation efforts and can help maintain adaptive potential (Capblancq et al. 2018; Capblancq and Forester 2021; Muñoz-Valencia et al. 2023). It is critical to consider multiple processes because genetic structure can be a result of colonization history, landscape connectivity, local adaptation, demography, or an interaction between processes (Orsini et al. 2013). Although genetic variance produced by each isolating process may not parse to unique drivers (Nadeau et al. 2016), RDA is a promising tool to identify what variance can or cannot be partitioned (Capblancq and Forester 2021). As the field of landscape genetics continues to develop, integrated approaches can guide conservation practices (Ruiz-Gonzalez et al. 2015; Priadka et al. 2019) and may prevent inflated correlations that can emerge if only a single process is considered.
References
Adamack AT, Gruber B (2014) PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol Evol 5:384–387
Agapow PM, Burt A (2001) Indices of multilocus linkage disequilibrium. Mol Ecol Notes 1:101–102
Alexander N (2023) Conservation of a fossorial grassland species (Geomys bursarius) through understanding niche reduction, landscape genetics, and phylogenetics. Dissertation, University of Illinois
Alexander N, Cosentino BJ, Schooley RL (2022) Testing the niche reduction hypothesis for a fossorial rodent (Geomys bursarius) experiencing agricultural intensification. Ecol Evol 12(ece3):9559
Alexander NB, Statham MJ, Sacks BN, Bean WT (2019) Generalist dispersal and gene flow of an endangered keystone specialist (Dipodomys ingens). J Mamm 100:1533–1545
Andersen DC (1987) Geomys bursarius burrowing patterns: influence of season and food patch structure. Ecology 68:1306–1318
Anderson CD, Epperson BK, Fortin MJ et al (2010) Considering spatial and temporal scale in landscape-genetic studies of gene flow. Mol Ecol. https://doi.org/10.1111/j.1365-294X.2010.04757.x
Augustine D, Davidson A, Dickinson K, Van Pelt B (2021) Thinking like a grassland: challenges and opportunities for biodiversity conservation in the Great Plains of North America. Rangel Ecol Manag 78:281–295
Austrich A, Mora MS, Mapelli FJ et al (2020) Influences of landscape characteristics and historical barriers on the population genetic structure in the endangered sand-dune subterranean rodent Ctenomys australis. Genetica 148:149–164
Barbosa S, Andrews KR, Goldberg AR et al (2021) The role of neutral and adaptive genomic variation in population diversification and speciation in two ground squirrel species of conservation concern. Mol Ecol 30:4673–4694
Borcard D, Legendre P (2002) All-scale spatial analysis of ecological data by means of principal coordinates of neighbour matrices. Ecol Modell 153:51–68
Bowcock AM, Ruiz-Linares A, Tomfohrde J et al (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457
Brookfield JFY (1996) A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol 5:453–544
Brown AHD, Feldmanz MW, Nevo E (1980) Multilocus structure of natural populations of Hordeum spontaneum. Genetics 96:523–536
Burbrink FT, Bernstein JM, Kuhn A et al (2022) Ecological divergence and the history of gene flow in the nearctic milksnakes (Lampropeltis triangulum complex). Syst Biol 71:839–858
Capblancq T, Forester BR (2021) Redundancy analysis: a Swiss Army Knife for landscape genomics. Methods Ecol Evol 12:2298–2309
Capblancq T, Luu K, Blum MGB, Bazin E (2018) Evaluation of redundancy analysis to identify signatures of local adaptation. Mol Ecol Resour 18:1223–1233
Centeno-Cuadros A, Romàn J, Delibes M, Godoy JA (2011) Prisoners in their habitat? Generalist dispersal by habitat specialists: a case study in southern water vole (Arvicola sapidus). PLoS ONE 6:e24613
Chen C, Durand E, Forbes F, Franҫois O (2007) Bayesian clustering algorithms ascertaining spatial population structure: a new computer program and a comparison study. Mol Ecol Notes 7:747–756
Connior MB (2011) Geomys bursarius (Rodentia: Geomyidae). Mamm Species 43:104–117
Cosentino BJ, Schooley RL, Bestelmeyer BT, McCarthy AJ, Sierzega K (2015) Rapid genetic restoration of a keystone species exhibiting delayed demographic response. Mol Ecol 24:6120–6133
Cushman SA, Landguth EL (2010) Spurious correlations and inference in landscape genetics. Mol Ecol 19:3592–3602
Cutrera AP, Vassallo AI, Mora MS et al (2013) Phylogeography and population genetic structure of the Talas tuco-tuco (Ctenomys talarum): integrating demographic and habitat histories. J Mamm. https://doi.org/10.1644/11-mamm-a-242.1
Dabrowski MJ, Pilot M, Kruczyk M et al (2014) Reliability assessment of null allele detection: inconsistencies between and within different methods. Mol Ecol 14:361–373
Daly JC, Patton JL (1990) Dispersal, gene flow, and allelic diversity between local populations of Thomomys bottae pocket gophers in the coastal ranges of California. Evolution 44:1283–1294
Davis WB, Ramsey RR, Arendale JM (1938) Distribution of pocket gophers (Geomys breviceps) in relation to soils. J Mamm 19:412–418
DeWoody JA, Harder AM, Mathur S, Willoughby JR (2021) The long-standing significance of genetic diversity in conservation. Mol Ecol 30:4147–4154
Donald McKay E, Harn AD, Follmer LR et al (1986) Wisconsinan and Sangamonian type sections of central Illinois. University of Illinois, Urbana-Champaign
Dray S, Dufour A-B (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20
Dobos RR, Kinast-Brown S, Roecker S, Lindbo DL (2023) Sandy soils in the United States: properties and use. In: Hartemink AE, Huang J (eds) Sandy soils. Progress in soil science. Springer, Cham, pp 25–38
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Elrod DA, Zimmerman EG, Sudman PD, Heidt GA (2000) A new subspecies of pocket gopher (genus Geomys) from the Ozark Mountains of Arkansas with comments on its historical biogeography. J Mamm 81:852–864
Epps CW, Keyghobadi N (2015) Landscape genetics in a changing world: disentangling historical and contemporary influences and inferring change. Mol Ecol 24:6021–6040
Esperandio IB, Ascensão F, Kindel A et al (2019) Do roads act as a barrier to gene flow of subterranean small mammals? A case study with Ctenomys minutus. Conserv Genet 20:385–393
Esri, HERE, Garmin, FAO, NOAA, USGS, © OpenStreetMap contributors, and the GIS User Community. 2017. World Topography Map. https://basemaps.arcgis.com/arcgis/rest/services/World_Basemap_v2/VectorTileServer. Accessed 5 May 2023
Esri, National Atlas of the United States, U.S. Geological Survey (USGS) (2010) U.S. major rivers file geodatabase feature class. https://www.arcgis.com/home/item.html?id=8206e517c2264bb39b4a0780462d5be1. Accessed 30 Apr 2023
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fasanella M, Bruno C, Cardoso Y, Lizarralde M (2013) Historical demography and spatial genetic structure of the subterranean rodent Ctenomys magellanicus in Tierra del Fuego (Argentina). Zool J Linn Soc 169:697–710
Galpern P, Peres-Neto PR, Polfus J, Manseau M (2014) MEMGENE: spatial pattern detection in genetic distance data. Methods Ecol Evol 5:1116–1120
Gilby BL, Olds AD, Brown CJ et al (2021) Applying systematic conservation planning to improve the allocation of restoration actions at multiple spatial scales. Restor Ecol 29:e13403
Genoways HH, Hamilton MJ, Bell DM et al (2008) Hybrid zones, genetic isolation, and systematics of pocket gophers (genus Geomys) in Nebraska. J Mamm 89:826–836
Gómez Fernández MJ, Boston ESM, Gaggiotti OE et al (2016) Influence of environmental heterogeneity on the distribution and persistence of a subterranean rodent in a highly unstable landscape. Genetica 144:711–722
Goudet J (2005) HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186
Hauser SS, Athrey G, Leberg PL (2021) Waste not, want not: Microsatellites remain an economical and informative technology for conservation genetics. Ecol Evol 11:15800–15814
Hedrick BP, Dickson BV, Dumont ER, Pierce SE (2020) The evolutionary diversity of locomotor innovation in rodents is not linked to proximal limb morphology. Sci Rep 10:717
Hendricksen RL (1972) Variation in the plains pocket gopher (Geomys bursarius) along a transect across Kansas and Eastern Colorado. Trans Kans Acad Sci 75:322
Hewitt GM (2004) The structure of biodiversity - Insights from molecular phylogeography. Front Zool 1:1–4
Hickman GC (1977) Swimming behavior in representative species of the three genera of North American geomyids. Southwest Nat 21:531–538
Hoffman JD, Choate JR (2008) Distribution and status of the yellow-faced pocket gopher in Kansas. Am Midl Nat 68:483–492
Hoffman JD, Choate JR, Channell R (2007) Effects of land use and soil texture on distribution of pocket gophers in Kansas. Southwest Nat 52:296–301
Hoffmeister DF (1989) Mammals of Illinois. University of Illinois, Chicago
Holderegger R, Wagner H (2008) Landscape genetics. Bioscience 58:199–207
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Illinois State Water Survey (2011). Major watersheds of Illinois. https://www.isws.illinois.edu/maps. Accessed 23 Mar 2023
Jenkins DG, Care M, Czerniewska J et al (2010) A meta-analysis of isolation by distance: relic or reference standard for landscape genetics? Ecography 33:316–320
Kamvar ZN, Tabima JF, Gr̈unwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014:1–14
Keller D, Holderegger R, Van Strien MJ (2013) Spatial scale affects landscape genetic analysis of a wetland grasshopper. Mol Ecol 22:2467–2482
Keller D, Holderegger R, van Strien MJ, Bolliger J (2015) How to make landscape genetics beneficial for conservation management? Conserv Genet 16:503–512
Kennerly TE (1963) Gene flow pattern and swimming ability of the pocket gopher. Southwest Nat 8:85–88
Kierepka EM, Latch EK (2016a) High gene flow in the American badger overrides habitat preferences and limits broadscale genetic structure. Mol Ecol 25:6055–6076
Kierepka EM, Latch EK (2016b) Fine-scale landscape genetics of the American badger (Taxidea taxus): Disentangling landscape effects and sampling artifacts in a poorly understood species. Heredity 116:33–43
Komarek EV, Spencer DA (1931) A new pocket gopher from Illinois and Indiana. J Mamm 12:404–408
Kopelman NM, Mayzel J, Jakobsson M et al (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191
Krupa JJ, Geluso KN (2000) Matching the color of excavated soil: cryptic coloration in the plains pocket gopher (Geomys bursarius). J Mammal 81:86–96
Lecis R, Dondina O, Orioli V et al (2022) Main roads and land cover shaped the genetic structure of a Mediterranean island wild boar population. Ecol Evol 12(ece3):8804
Lucati F, Poignet M, Miró A et al (2020) Multiple glacial refugia and contemporary dispersal shape the genetic structure of an endemic amphibian from the Pyrenees. Mol Ecol 29:2904–2921
Manel S, Holderegger R (2013) Ten years of landscape genetics. Trends Ecol Evol 28:614–621
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18:189–197
Mapelli FJ, Boston ESM, Fameli A et al (2020) Fragmenting fragments: landscape genetics of a subterranean rodent (Mammalia, Ctenomyidae) living in a human-impacted wetland. Landsc Ecol 35:1089–1106
Mapelli FJ, Mora MS, Mirol PM, Kittlein MJ (2012) Population structure and landscape genetics in the endangered subterranean rodent Ctenomys porteousi. Conserv Genet 13:165–181
Mauk CL, Houck MA, Bradley RD (1999) Morphometric analysis of seven species of pocket gophers (Geomys). J Mamm 80:499–511
McCluskey EM, Lulla V, Peterman WE et al (2022) Linking genetic structure, landscape genetics, and species distribution modeling for regional conservation of a threatened freshwater turtle. Landsc Ecol 37:1017–1034
Meeûs TD (2018) Revisiting FIS, FST, Wahlund effects, and null alleles. Heredity 109:446–456
Muñoz-Valencia V, Montoya-Lerma J, Seppä P, Diaz F (2023) Landscape genetics across the Andes mountains: Environmental variation drives genetic divergence in the leaf-cutting ant Atta cephalotes. Mol Ecol 32:95–109
Musher LJ, Giakoumis M, Albert J et al (2022) River network rearrangements promote speciation in lowland Amazonian birds. Sci Adv 8:eabn1099
Nadeau S, Meirmans PG, Aitken SN et al (2016) The challenge of separating signatures of local adaptation from those of isolation by distance and colonization history: the case of two white pines. Ecol Evol 6:8649–8664
Oksanen J, Simpson G, Blanchet F et al (2022) Vegan: community ecology package. R package version 2.6-4
Orsini L, Vanoverbeke J, Swillen I et al (2013) Drivers of population genetic differentiation in the wild: Isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Mol Ecol 22:5983–5999
Painter LE, Weldy MJ, Crowhurst RS et al (2022) Landscape genetics of the camas pocket gopher (Thomomys bulbivorus), an endemic mammal of Oregon’s Willamette Valley. West N Am Nat 82:479–493
Paradis E (2010) Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420
Penney DF, Zimmerman EG (1976) Genic divergence and local population differentiation by random drift in the pocket gopher genus Geomys. Evolution 30:473–483
Pfau RS, van den Bussche RA, McBee K (2001) Population genetics of the hispid cotton rat (Sigmodon hispidus): Patterns of genetic diversity at the major histocompatibility complex. Mol Ecol 10:1939–1945
Priadka P, Manseau M, Trottier T et al (2019) Partitioning drivers of spatial genetic variation for a continuously distributed population of boreal caribou: Implications for management unit delineation. Ecol Evol 9:141–153
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Puechmaille SJ (2016) The program structure does not reliably recover the correct population structure when sampling is uneven: Subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627
Pynne JT, Owens JM, Castleberry SB et al (2019) Movement and fate of translocated and in situ southeastern pocket gophers. Southeast Nat 18:297–302
Reichman OJ, Seabloom EW (2002) The role of pocket gophers as subterranean ecosystem engineers. Trends Ecol Evol 17:44–49
Richardson JL, Urban MC, Bolnick DI, Skelly DK (2014) Microgeographic adaptation and the spatial scale of evolution. Trends Ecol Evol 29:165–179
Rios E, Álvarez-Castañeda ST (2012) Pelage color variation in pocket gophers (Rodentia: Geomyidae) in relation to sex, age and differences in habitat. Mamm Biol 77:160–165
Roratto PA, Fernandes FA, de Freitas TRO (2015) Phylogeography of the subterranean rodent Ctenomys torquatus: an evaluation of the riverine barrier hypothesis. J Biogeogr 42:694–705
Ruiz-Gonzalez A, Cushman SA, Madeira MJ et al (2015) Isolation by distance, resistance and/or clusters? Lessons learned from a forest-dwelling carnivore inhabiting a heterogeneous landscape. Mol Ecol 24:5110–5129
Schulze DG, Nagel JL, Van Scoyoc GE et al (1993) Significance of organic matter in determining soil colors. Soil color. Wiley, New York, pp 71–90
Sexton JP, Hangartner SB, Hoffmann AA (2014) Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution 68:1–15
Shafer ABA, Wolf JBW (2013) Widespread evidence for incipient ecological speciation: a meta-analysis of isolation-by-ecology. Ecol Lett 16:940–950
Shirk AJ, Landguth EL, Cushman SA (2017) A comparison of individual-based genetic distance metrics for landscape genetics. Mol Ecol Resour 17:1308–1317
Sikes RS, The Animal Use and Care Committee of the American Society of Mammalogists (2016) Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J Mamm 92:235–253
Singaravelan N, Raz S, Tzur S et al (2013) Adaptation of pelage color and pigment variations in Israeli subterranean blind mole rats, Spalax ehrenbergi. PLoS ONE 8:e69346
Skey ED, Ottewell KM, Spencer PB et al (2023) Empirical landscape genetic comparison of single nucleotide polymorphisms and microsatellites in three arid-zone mammals with high dispersal capacity. Ecol Evol 13:e10037
Smith PW (1957) An analysis of post-Wisconsin biogeography of the Prairie Peninsula region based on distributional phenomena among terrestrial vertebrate populations. Ecology 38:205–218
Soil Survey Staff, Natural Resources Conservation Service, United States Department of Agriculture (2018) Soil survey geographic (STATSO2) database for United States. https://data.nal.usda.gov/dataset/us-general-soil-map-statsgo2-individual-states. Accessed 22 Jan 2018
Soil Survey Staff (2022) Gridded national soil survey geographic (gNATSGO) database for the conterminous United States. United States Department of Agriculture, Natural Resources Conservation Service. https://nrcs.app.box.com/v/soils. Accessed 27 Mar 2023
Steinberg EK (1999) Characterization of polymorphic microsatellites from current and historic populations of North American pocket gophers (Geomyidae: Thomomys). Mol Ecol 8:1075–1092
Sudman PD, Choate JR, Zimmerman EG (1987) Taxonomy of chromosomal races of Geomys bursarius lutescens Merriam. J Mamm 68:526–543
Vähä JP, Erkinaro J, Niemelä E, Primmer CR (2007) Life-history and habitat features influence the within-river genetic structure of Atlantic salmon. Mol Ecol 16:2638–2654
van Strien MJ, Holderegger R, van Heck HJ (2015) Isolation-by-distance in landscapes: considerations for landscape genetics. Heredity 114:27–37
Vaughan TA (1963) Movement made by two species of pocket gophers. Am Midl Nat 69:367–372
Visser JH, Bennett NC, Van Vuuren BJ (2018) Spatial genetic diversity in the Cape mole-rat, Georychus capensis: Extreme isolation of populations in a subterranean environment. PLoS ONE 13:e0194165
Vleck D (1981) Burrow structure and foraging costs in the fossorial rodent, Thomomys bottae. Oecologia 49:391–396
Vleck D (1979) The energy cost of burrowing by the pocket gopher Thomomys bottae. Physiol Zool 52:122–136
Wagner H (2022) 6.3 Worked example. In: Wagner H (ed) Landscape genetic data analysis in R. Toronto
Wang IJ, Bradburd GS (2014) Isolation by environment. Mol Ecol 23:5649–5662
Waraniak JM, Fisher JDL, Purcell K et al (2019) Landscape genetics reveal broad and fine-scale population structure due to landscape features and climate history in the northern leopard frog (Rana pipiens) in North Dakota. Ecol Evol 9:1041–1060
Warner RE (1994) Agricultural land use and grassland habitat in Illinois: future shock for midwestern birds? Conserv Biol 8:147–156
Warnock WG, Rasmussen JB, Taylor EB (2010) Genetic clustering methods reveal bull trout (salvelinus confluentus) fine-scale population structure as a spatially nested hierarchy. Conserv Genet 11:1421–1433
Warren AE, Conner LM, Castleberry SB, Markewitz D (2017) Home range, survival, and activity patterns of the southeastern pocket gopher: Implications for translocation. J Fish Wildl Manag 8:544–557
Wascher HL, Alexander JD, Ray BW et al (1960) Characteristics of soils associated with glacial tills in northeastern Illinois. University of Illinois, Urbana
Weber JN, Bradburd GS, Stuart YE et al (2016) Partitioning the effects of isolation by distance, environment, and physical barriers on genomic divergence between parapatric threespine stickleback. Evolution 71:342–356
Weir BS, Cokerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Welborn SR, Light JE (2014) Population genetic structure of the baird’s pocket gopher, Geomys breviceps, in Eastern Texas. West N Am Nat 74:325–334
Welborn SR, Renshaw MA, Light JE (2012) Characterization of 10 polymorphic loci in the Baird’s pocket gopher (Geomys breviceps) and cross-amplification in other gopher species. Conserv Genet Resour 4:467–469
Wilkins KT, Roberts HR (2007) Comparative analysis of burrow systems of seven species of pocket gophers (Rodentia: Geomyidae). Southwest Nat 52:83–88
Wilkins KT, Swearingen CD (1990) Factors affecting historical distribution and modern geographic variation in the South Texas pocket gopher Geomys personatus. Am Midl Nat 124:57–72
Williams LR, Cameron GN (1984) Demography of dispersal in Attwater’s pocket gopher (Geomys attwateri). J Mammal 65:67–75
Wlasiuk G, Nachman MW (2007) The genetics of adaptive coat color in gophers: coding variation at Mc1r is not responsible for dorsal color differences. J Hered 98:567–574
Wright S (1943) Isolation by distance. Genetics 28:114–138
Acknowledgements
Our research was funded by the USFWS/IDNR Federal Aid in Wildlife Restoration Program (project number W-191-R-1). We thank B. Bluett and S. McTaggart for project administration and field surveys. We thank J. Merrit and J. Mengelkoch from the Illinois Natural History Survey for providing specimens for genetic samples. We are grateful to J. Light for providing input on DNA extraction and DNA samples for positive controls. We also thank S. Beyer, W. Berry, and K. Fountain for fieldwork assistance, and B. Otaibi and A. Bryan for assistance with DNA extraction. E. Larson, J. Fraterrigo, K. Andreoni, A. Cervantes, and C. Wagnon provided valuable discussion and review of our work and this manuscript.
Funding
This work was supported by the USFWS/IDNR Federal Aid in Wildlife Restoration Program (Project Number W-191-R-1).
Author information
Authors and Affiliations
Contributions
NA wrote the main manuscript text and prepared figures and tables. NA, RLS, and BJC made substantial contributions to the conception and the design of the work, drafted or revised the manuscript with significant intellectual contributions, and approved the version to be published. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests, financial or otherwise, to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alexander, N., Cosentino, B.J. & Schooley, R.L. Partitioning genetic structure of a subterranean rodent at multiple spatial scales: accounting for isolation by barriers, distance, and environment. Landsc Ecol 39, 92 (2024). https://doi.org/10.1007/s10980-024-01878-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10980-024-01878-0